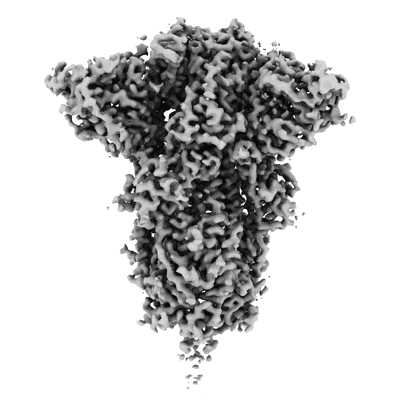
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7s6i
Q-score: 0.514
Torres JL, Ozorowski G, Andreano E, Liu H, Copps J, Piccini G, Donnici L, Conti M, Planchais C, Planas D, Manganaro N, Pantano E, Paciello I, Pileri P, Bruel T, Montomoli E, Mouquet H, Schwartz O, Sala C, De Francesco R, Wilson IA, Rappuoli R, Ward AB
PNAS (2022) 119 pp. e2120976119-e2120976119 [ DOI: doi:10.1073/pnas.2120976119 Pubmed: 35549549 ]
J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (conformation 3)
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7s6j
Q-score: 0.376
Torres JL, Ozorowski G, Andreano E, Liu H, Copps J, Piccini G, Donnici L, Conti M, Planchais C, Planas D, Manganaro N, Pantano E, Paciello I, Pileri P, Bruel T, Montomoli E, Mouquet H, Schwartz O, Sala C, De Francesco R, Wilson IA, Rappuoli R, Ward AB
PNAS (2022) 119 pp. e2120976119-e2120976119 [ DOI: doi:10.1073/pnas.2120976119 Pubmed: 35549549 ]
- J08 fragment antigen binding heavy chain variable domain (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- J08 fragment antigen binding light chain variable domain (11 kDa, Protein from Homo sapiens)
- J08 fragment antigen binding in complex with sars-2-cov-6p-mut2 s protein (690 kDa, Complex)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 2)
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7s6k
Q-score: 0.416
Torres JL, Ozorowski G, Andreano E, Liu H, Copps J, Piccini G, Donnici L, Conti M, Planchais C, Planas D, Manganaro N, Pantano E, Paciello I, Pileri P, Bruel T, Montomoli E, Mouquet H, Schwartz O, Sala C, De Francesco R, Wilson IA, Rappuoli R, Ward AB
PNAS (2022) 119 pp. e2120976119-e2120976119 [ DOI: doi:10.1073/pnas.2120976119 Pubmed: 35549549 ]
- J08 fragment antigen binding heavy chain variable domain (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- J08 fragment antigen binding light chain variable domain (11 kDa, Protein from Homo sapiens)
- J08 fragment antigen binding in complex with sars-2-cov-6p-mut2 s protein (690 kDa, Complex)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (conformation 3)
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 7s6l
Q-score: 0.299
Torres JL, Ozorowski G, Andreano E, Liu H, Copps J, Piccini G, Donnici L, Conti M, Planchais C, Planas D, Manganaro N, Pantano E, Paciello I, Pileri P, Bruel T, Montomoli E, Mouquet H, Schwartz O, Sala C, De Francesco R, Wilson IA, Rappuoli R, Ward AB
PNAS (2022) 119 pp. e2120976119-e2120976119 [ Pubmed: 35549549 DOI: doi:10.1073/pnas.2120976119 ]
- J08 fragment antigen binding in complex with sars-2-cov-6p-mut7 s protein (590 kDa, Complex)
- J08 fragment antigen binding light chain variable domain (11 kDa, Protein from Homo sapiens)
- J08 fragment antigen binding heavy chain variable domain (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
J08 fragment antigen binding in complex with Omicron SARS-CoV-2-6P S protein
Torres JL, Ozorowski G, Andreano E, Liu H, Copps J, Piccini G, Donnici L, Conti M, Planchais C, Planas D, Manganaro N, Pantano E, Paciello I, Pileri P, Bruel T, Montomoli E, Mouquet H, Schwartz O, Sala C, De Francesco R, Wilson IA, Rappuoli R, Ward AB
PNAS (2022) 119 pp. e2120976119-e2120976119 [ DOI: doi:10.1073/pnas.2120976119 Pubmed: 35549549 ]
- J08 fragment antigen binding light chain variable domain (Macromolecule from Homo sapiens)
- J08 fragment antigen binding heavy chain variable domain (Macromolecule from Homo sapiens)
- Omicron sars-cov-2-6p s protein (Macromolecule from Severe acute respiratory syndrome coronavirus 2)
- J08 fragment antigen binding in complex with omicron sars-cov-2-6p s protein (Complex from Severe acute respiratory syndrome coronavirus 2)
SARS-2 CoV 6P Mut7 in complex with Fab J08
Andreano E, Nicastri E, Paciello I, Pileri P, Manganaro N, Piccini G, Manenti A, Pantano E, Kabanova A, Troisi M, Vacca F, Cardamone D, De Santi C, Torres JL, Ozorowski G, Benincasa L, Jang H, Di Genova C, Depau L, Brunetti J, Agrati C, Capobianchi MR, Castilleti C, Emiliozzi A, Fabianni M, Montagnani F, Bracci L, Sautto G, Ross T, Mantomoli E, Temperton N, Ward AB, Sala C, Ippolito G, Rappuoli R
To Be Published
SARS-2 CoV 6P Mut7 in complex with Fab F05
Andreano E, Nicastri E, Paciello I, Pileri P, Manganaro N, Piccini G, Manenti A, Pantano E, Kabanova A, Troisi M, Vacca F, Cardamone D, De Santi C, Torres JL, Ozorowski G, Benincasa L, Jang H, Di Genova C, Depau L, Brunetti J, Agrati C, Capobianchi MR, Castilleti C, Emiliozzi A, Fabianni M, Montagnani F, Bracci L, Sautto G, Ross T, Mantomoli E, Temperton N, Ward AB, Sala C, Ippolito G, Rappuoli R
To Be Published
SARS-2 CoV 6P Mut 7 in complex with fragment antigen binding (fab) I14
Andreano E, Nicastri E, Paciello I, Pileri P, Manganaro N, Piccini G, Manenti A, Pantano E, Kabanova A, Troisi M, Vacca F, Cardamone D, De Santi C, Torres JL, Ozorowski G, Benincasa L, Jang H, Di Genova C, Depau L, Brunetti J, Agrati C, Capobianchi MR, Castilleti C, Emiliozzi A, Fabianni M, Montagnani F, Bracci L, Sautto G, Ross T, Mantomoli E, Temperton N, Ward AB, Sala C, Ippolito G, Rappuoli R
To Be Published