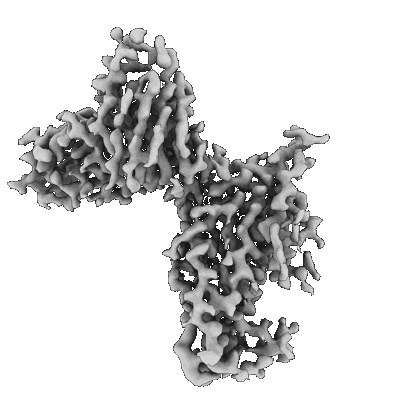
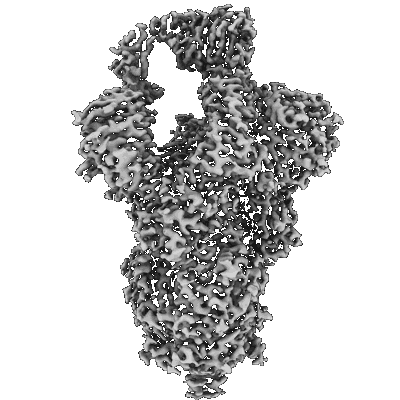
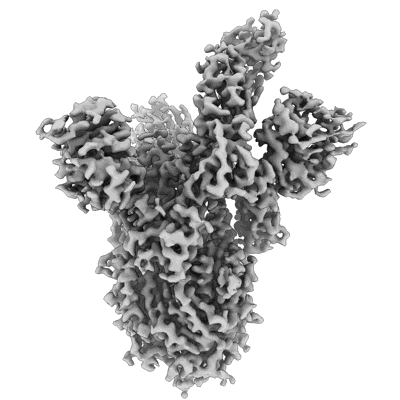
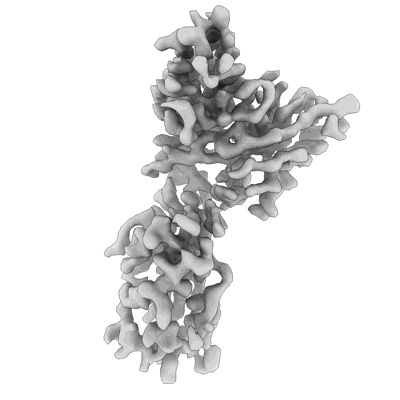Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8erq
Q-score: 0.493
Park YJ, Pinto D, Walls AC, Liu Z, De Marco A, Benigni F, Zatta F, Silacci-Fregni C, Bassi J, Sprouse KR, Addetia A, Bowen JE, Stewart C, Giurdanella M, Saliba C, Guarino B, Schmid MA, Franko NM, Logue JK, Dang HV, Hauser K, di Iulio J, Rivera W, Schnell G, Rajesh A, Zhou J, Farhat N, Kaiser H, Montiel-Ruiz M, Noack J, Lempp FA, Janer J, Abdelnabi R, Maes P, Ferrari P, Ceschi A, Giannini O, de Melo GD, Kergoat L, Bourhy H, Neyts J, Soriaga L, Purcell LA, Snell G, Whelan SPJ, Lanzavecchia A, Virgin HW, Piccoli L, Chu HY, Pizzuto MS, Corti D, Veesler D
Science (2022) 378 pp. 619-627 [ Pubmed: 36264829 DOI: doi:10.1126/science.adc9127 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 ba.1 spike ectodomain trimer in complex with the s2x324 neutralizing antibody fab fragment (local refinement of the rbd and s2x324) (Complex from Severe acute respiratory syndrome coronavirus 2)
- S2x324 fab heavy chain (13 kDa, Protein from Homo sapiens)
- Sars-cov-2 omicron ba.1 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- S2x324 fab (Complex from Homo sapiens)
- S2x324 fab light chain (11 kDa, Protein from Homo sapiens)
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8err
Q-score: 0.449
Park YJ, Pinto D, Walls AC, Liu Z, De Marco A, Benigni F, Zatta F, Silacci-Fregni C, Bassi J, Sprouse KR, Addetia A, Bowen JE, Stewart C, Giurdanella M, Saliba C, Guarino B, Schmid MA, Franko NM, Logue JK, Dang HV, Hauser K, di Iulio J, Rivera W, Schnell G, Rajesh A, Zhou J, Farhat N, Kaiser H, Montiel-Ruiz M, Noack J, Lempp FA, Janer J, Abdelnabi R, Maes P, Ferrari P, Ceschi A, Giannini O, de Melo GD, Kergoat L, Bourhy H, Neyts J, Soriaga L, Purcell LA, Snell G, Whelan SPJ, Lanzavecchia A, Virgin HW, Piccoli L, Chu HY, Pizzuto MS, Corti D, Veesler D
Science (2022) 378 pp. 619-627 [ Pubmed: 36264829 DOI: doi:10.1126/science.adc9127 ]
- S2x324 heavy chain (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- S2x324 light chain (11 kDa, Protein from Homo sapiens)
- Sars-cov-2 omicron ba.1 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 omicron ba.1 spike ectodomain trimer in complex with the s2x324 neutralizing antibody fab fragment (Complex from Severe acute respiratory syndrome coronavirus 2)
- S2x324 fab (Complex from Homo sapiens)
Resolution: 3.95 Å
EM Method: Single-particle
Fitted PDBs: 7r7n
Q-score: 0.411
Starr TN, Czudnochowski N, Liu Z, Zatta F, Park YJ, Addetia A, Pinto D, Beltramello M, Hernandez P, Greaney AJ, Marzi R, Glass WG, Zhang I, Dingens AS, Bowen JE, Tortorici MA, Walls AC, Wojcechowskyj JA, De Marco A, Rosen LE, Zhou J, Montiel-Ruiz M, Kaiser H, Dillen JR, Tucker H, Bassi J, Silacci-Fregni C, Housley MP, di Iulio J, Lombardo G, Agostini M, Sprugasci N, Culap K, Jaconi S, Meury M, Dellota Jr E, Abdelnabi R, Foo SC, Cameroni E, Stumpf S, Croll TI, Nix JC, Havenar-Daughton C, Piccoli L, Benigni F, Neyts J, Telenti A, Lempp FA, Pizzuto MS, Chodera JD, Hebner CM, Virgin HW, Whelan SPJ, Veesler D, Corti D, Bloom JD, Snell G
Nature (2021) 597 pp. 97-102 [ DOI: doi:10.1038/s41586-021-03807-6 Pubmed: 34261126 ]
- S2d106 fab heavy chain (13 kDa, Protein from Homo sapiens)
- S2d106 fab light chain (11 kDa, Protein from Homo sapiens)
- S2d106 (Complex from Homo sapiens)
- Sars-cov-2 spike in complex with the s2d106 neutralizing antibody fab fragment (Complex)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 spike in complex with the S2D106 neutralizing antibody Fab fragment
Starr TN, Czudnochowski N, Liu Z, Zatta F, Park YJ, Addetia A, Pinto D, Beltramello M, Hernandez P, Greaney AJ, Marzi R, Glass WG, Zhang I, Dingens AS, Bowen JE, Tortorici MA, Walls AC, Wojcechowskyj JA, De Marco A, Rosen LE, Zhou J, Montiel-Ruiz M, Kaiser H, Dillen JR, Tucker H, Bassi J, Silacci-Fregni C, Housley MP, di Iulio J, Lombardo G, Agostini M, Sprugasci N, Culap K, Jaconi S, Meury M, Dellota Jr E, Abdelnabi R, Foo SC, Cameroni E, Stumpf S, Croll TI, Nix JC, Havenar-Daughton C, Piccoli L, Benigni F, Neyts J, Telenti A, Lempp FA, Pizzuto MS, Chodera JD, Hebner CM, Virgin HW, Whelan SPJ, Veesler D, Corti D, Bloom JD, Snell G
Nature (2021) 597 pp. 97-102 [ DOI: doi:10.1038/s41586-021-03807-6 Pubmed: 34261126 ]
Starr TN, Czudnochowski N, Liu Z, Zatta F, Park YJ, Addetia A, Pinto D, Beltramello M, Hernandez P, Greaney AJ, Marzi R, Glass WG, Zhang I, Dingens AS, Bowen JE, Tortorici MA, Walls AC, Wojcechowskyj JA, De Marco A, Rosen LE, Zhou J, Montiel-Ruiz M, Kaiser H, Dillen JR, Tucker H, Bassi J, Silacci-Fregni C, Housley MP, di Iulio J, Lombardo G, Agostini M, Sprugasci N, Culap K, Jaconi S, Meury M, Dellota Jr E, Abdelnabi R, Foo SC, Cameroni E, Stumpf S, Croll TI, Nix JC, Havenar-Daughton C, Piccoli L, Benigni F, Neyts J, Telenti A, Lempp FA, Pizzuto MS, Chodera JD, Hebner CM, Virgin HW, Whelan SPJ, Veesler D, Corti D, Bloom JD, Snell G
Nature (2021) 597 pp. 97-102 [ DOI: doi:10.1038/s41586-021-03807-6 Pubmed: 34261126 ]