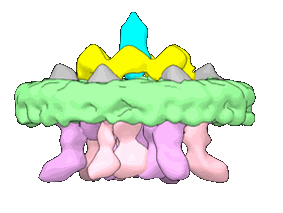
penton capsomer of the VZV C-capsid
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 8xa0
Q-score: 0.195
Cao L, Wang N, Lv Z, Chen W, Chen Z, Song L, Sha X, Wang G, Hu Y, Lian X, Cui G, Fan J, Quan Y, Liu H, Hou H, Wang X
Hlife (2024) 2 pp. 64-74 [ DOI: doi:10.1016/j.hlife.2023.10.007 ]
- Capsid vertex component 1 (73 kDa, Protein from Human alphaherpesvirus 3)
- Large tegument protein deneddylase (5 kDa, Protein from Human alphaherpesvirus 3)
- Tri1 (32 kDa, Protein from Human alphaherpesvirus 3)
- Human alphaherpesvirus 3 (Virus from Human alphaherpesvirus 3)
- Capsid vertex component 2 (10 kDa, Protein from Human alphaherpesvirus 3)
- Tri2b (28 kDa, Protein from Human alphaherpesvirus 3)
- Tri2a (27 kDa, Protein from Human alphaherpesvirus 3)
- Major capsid protein (152 kDa, Protein from Human alphaherpesvirus 3)
Penton capsomer of the VZV B-Capsid
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 8xa2
Q-score: 0.217
Cao L, Wang N, Lv Z, Chen W, Chen Z, Song L, Sha X, Wang G, Hu Y, Lian X, Cui G, Fan J, Quan Y, Liu H, Hou H, Wang X
Hlife (2024) 2 pp. 64-74 [ DOI: doi:10.1016/j.hlife.2023.10.007 ]
- Tri1 (32 kDa, Protein from Human alphaherpesvirus 3)
- Human alphaherpesvirus 3 (Virus from Human alphaherpesvirus 3)
- Tri2b (28 kDa, Protein from Human alphaherpesvirus 3)
- Major capsid protein (152 kDa, Protein from Human alphaherpesvirus 3)
- Tri2a (27 kDa, Protein from Human alphaherpesvirus 3)
CryoEM structure of the TIR domain from AbTir in complex with 3AD
Resolution: 2.74 Å
EM Method: Helical reconstruction
Fitted PDBs: 7uxu
Q-score: 0.636
Manik MK, Shi Y, Li S, Zaydman MA, Damaraju N, Eastman S, Smith TG, Gu W, Masic V, Mosaiab T, Weagley JS, Hancock SJ, Vasquez E, Hartley-Tassell L, Kargios N, Maruta N, Lim BYJ, Burdett H, Landsberg MJ, Schembri MA, Prokes I, Song L, Grant M, DiAntonio A, Nanson JD, Guo M, Milbrandt J, Ve T, Kobe B
Science (2022) 377 pp. eadc8969-eadc8969 [ Pubmed: 36048923 DOI: doi:10.1126/science.adc8969 ]
- Molecular chaperone tir (15 kDa, Protein from Acinetobacter baumannii)
- [[(2~{r},3~{s},4~{r},5~{r})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{r},3~{s},4~{r},5~{r})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate (686 Da, Ligand)
- Filament of the abtir tir domain in complex with 3ad (Complex from Acinetobacter baumannii)
Porphyromonas gingivalis type IX secretion system
Song L, Perpich JD, Wu C, Doan T, Nowakowska Z, Potempa J, Christie PJ, Cascales E, Lamont RJ, Hu B
PNAS (2022) 119 pp. e2119907119-e2119907119 [ Pubmed: 35471908 DOI: doi:10.1073/pnas.2119907119 ]
- T9ss (Complex from Porphyromonas gingivalis)
Song L, Perpich JD, Wu C, Doan T, Nowakowska Z, Potempa J, Christie PJ, Cascales E, Lamont RJ, Hu B
PNAS (2022) 119 pp. e2119907119-e2119907119 [ Pubmed: 35471908 DOI: doi:10.1073/pnas.2119907119 ]
- T9ss (Complex from Porphyromonas gingivalis)
Song L, Perpich JD, Wu C, Doan T, Nowakowska Z, Potempa J, Christie PJ, Cascales E, Lamont RJ, Hu B
PNAS (2022) 119 pp. e2119907119-e2119907119 [ Pubmed: 35471908 DOI: doi:10.1073/pnas.2119907119 ]
- T9ss (Complex from Porphyromonas gingivalis)
pKM101-T4SS outer membrane core complex
Khara P, Song L, Christie PJ, Hu B
Mbio (2021) 12 pp. e0246521-e0246521 [ DOI: doi:10.1128/mBio.02465-21 Pubmed: 34634937 ]
pKM101-T4SS inner membrane complex
Khara P, Song L, Christie PJ, Hu B
Mbio (2021) 12 pp. e0246521-e0246521 [ DOI: doi:10.1128/mBio.02465-21 Pubmed: 34634937 ]
SARS-CoV-2 S-6P in complex with 9 Fabs
Resolution: 3.16 Å
EM Method: Single-particle
Fitted PDBs: 7e8c
Q-score: 0.39
Cao Y, Yisimayi A, Bai Y, Huang W, Li X, Zhang Z, Yuan T, An R, Wang J, Xiao T, Du S, Ma W, Song L, Li Y, Li X, Song W, Wu J, Liu S, Li X, Zhang Y, Su B, Guo X, Wei Y, Gao C, Zhang N, Zhang Y, Dou Y, Xu X, Shi R, Lu B, Jin R, Ma Y, Qin C, Wang Y, Feng Y, Xiao J, Xie XS
Cell Res (2021) 31 pp. 732-741 [ DOI: doi:10.1038/s41422-021-00514-9 Pubmed: 34021265 ]
- 604 h (23 kDa, Protein from Homo sapiens)
- N9 l (22 kDa, Protein from Homo sapiens)
- N9 h (24 kDa, Protein from Homo sapiens)
- 604 l (23 kDa, Protein from Homo sapiens)
- Sars-cov-2 s (Complex from Homo sapiens)
- 368-2 h (24 kDa, Protein from Homo sapiens)
- Sars-cov-2 s-6p in complex with 9 fabs (Complex from Severe acute respiratory syndrome coronavirus 2)
- 604 h, 604 l, n9 h, 368-2 l, 368-2 h, n9 l (Complex)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 368-2 l (23 kDa, Protein from Homo sapiens)
SARS-CoV-2 NTD in complex with N9 Fab
Resolution: 3.18 Å
EM Method: Single-particle
Fitted PDBs: 7e8f
Q-score: 0.592
Cao Y, Yisimayi A, Bai Y, Huang W, Li X, Zhang Z, Yuan T, An R, Wang J, Xiao T, Du S, Ma W, Song L, Li Y, Li X, Song W, Wu J, Liu S, Li X, Zhang Y, Su B, Guo X, Wei Y, Gao C, Zhang N, Zhang Y, Dou Y, Xu X, Shi R, Lu B, Jin R, Ma Y, Qin C, Wang Y, Feng Y, Xiao J, Xie XS
Cell Res (2021) 31 pp. 732-741 [ Pubmed: 34021265 DOI: doi:10.1038/s41422-021-00514-9 ]
- Sars-cov-2 ntd in complex with n9 fab (Complex from SARS coronavirus Tor2)
- 368-2 l (23 kDa, Protein from Homo sapiens)
- Spike protein s1 (25 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- N9 l (22 kDa, Protein from Homo sapiens)
- Spike protein s1 (33 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 604 l (23 kDa, Protein from Homo sapiens)
- 368-2 h (24 kDa, Protein from Homo sapiens)
- Sars-cov-2 ntd and rbd (Complex from Homo sapiens)
- N9 h (24 kDa, Protein from Homo sapiens)
- 604 h (23 kDa, Protein from Homo sapiens)
- N9, 368-2 and 604h fab (Complex)