Resolution: 2.79 Å
EM Method: Single-particle
Fitted PDBs: 8vxh
Q-score: 0.573
Ashraf KU, Bunoro-Batista M, Ansell TB, Punetha A, Rosario-Garrido S, Firlar E, Kaelber JT, Stansfeld PJ, Petrou VI
bioRxiv (2025) [ DOI: doi:10.1101/2025.01.29.634835 ]
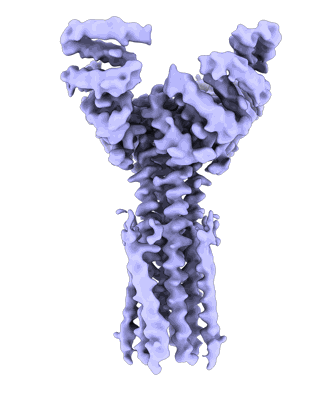
- Undecaprenyl-phosphate 4-deoxy-4-formamido-l-arabinose transferase (40 kDa, Protein from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2)
- Salmonella enterica arnc in msp1e3d1 nanodisc in the apo state (162 kDa, Complex from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2)
Resolution: 2.96 Å
EM Method: Single-particle
Fitted PDBs: 9asc
Q-score: 0.5
Ashraf KU, Bunoro-Batista M, Ansell TB, Punetha A, Rosario-Garrido S, Firlar E, Kaelber JT, Stansfeld PJ, Petrou VI
bioRxiv (2025) [ DOI: doi:10.1101/2025.01.29.634835 ]
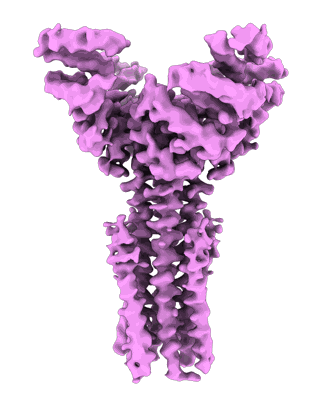
- Uridine-5'-diphosphate (404 Da, Ligand)
- Salmonella enterica arnc in msp1e3d1 nanodisc bound to udp and mn2+ (162 kDa, Complex from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2)
- Undecaprenyl-phosphate 4-deoxy-4-formamido-l-arabinose transferase (40 kDa, Protein from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2)
- Manganese (ii) ion (54 Da, Ligand)
Resolution: 2.74 Å
EM Method: Single-particle
Fitted PDBs: 9b77
Q-score: 0.599
Ashraf KU, Bunoro-Batista M, Ansell TB, Punetha A, Rosario-Garrido S, Firlar E, Kaelber JT, Stansfeld PJ, Petrou VI
bioRxiv (2025) [ DOI: doi:10.1101/2025.01.29.634835 ]
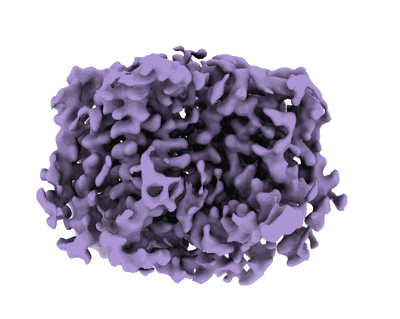
- Undecaprenyl-phosphate 4-deoxy-4-formamido-l-arabinose transferase (40 kDa, Protein from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2)
- Water (18 Da, Ligand)
- Salmonella enterica arnc in msp1e3d1 nanodisc in the apo state (162 kDa, Complex from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2)
Structure of the MlaCD complex (1:6 stoichiometry)
Resolution: 4.35 Å
EM Method: Single-particle
Fitted PDBs: 8oj4
Q-score: 0.448
Wotherspoon P, Johnston H, Hardy DJ, Holyfield R, Bui S, Ratkeviciute G, Sridhar P, Colburn J, Wilson CB, Colyer A, Cooper BF, Bryant JA, Hughes GW, Stansfeld PJ, Bergeron JRC, Knowles TJ
Nat Commun (2024) 15 pp. 6394-6394 [ Pubmed: 39080293 DOI: doi:10.1038/s41467-024-50615-3 ]
Structure of the MlaCD complex (2:6 stoichiometry)
Resolution: 4.38 Å
EM Method: Single-particle
Fitted PDBs: 8ojg
Q-score: 0.421
Wotherspoon P, Johnston H, Hardy DJ, Holyfield R, Bui S, Ratkeviciute G, Sridhar P, Colburn J, Wilson CB, Colyer A, Cooper BF, Bryant JA, Hughes GW, Stansfeld PJ, Bergeron JRC, Knowles TJ
Nat Commun (2024) 15 pp. 6394-6394 [ Pubmed: 39080293 DOI: doi:10.1038/s41467-024-50615-3 ]
Artificial membrane protein TMHC4_R (ROCKET)
Abramsson M, Corey RA, Skerle J, Persson L, Anden O, Oluwole AO, Howard RJ, Lindahl E, Robinson CV, Strisovsky K, Marklund EG, Drew D, Stansfeld PJ, Landreh M
To Be Published
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 8tlu
Q-score: 0.434
Marmont LS, Orta AK, Baileeves BWA, Sychantha D, Fernandez-Galliano A, Li YE, Greene NG, Corey RA, Stansfeld PJ, Clemons Jr WM, Bernhardt TG
Nat Microbiol (2024) 9 pp. 763-775 [ DOI: doi:10.1038/s41564-024-01603-2 Pubmed: 38336881 ]
- Phospho-n-acetylmuramoyl-pentapeptide-transferase (39 kDa, Protein from Escherichia coli K-12)
- E. coli mray t23p dimer (39 kDa, Cellular component from Escherichia coli K-12)
In situ structure of the Nitrosopumilus maritimus S-layer - Six-fold symmetry (C6)
Resolution: 3.4 Å
EM Method: Subtomogram averaging
Fitted PDBs: 8c8o
Q-score: 0.434
von Kugelgen A, Cassidy CK, van Dorst S, Pagani LL, Batters C, Ford Z, Lowe J, Alva V, Stansfeld PJ, Bharat TAM
Nature (2024) 630 pp. 230-236 [ Pubmed: 38811725 DOI: doi:10.1038/s41586-024-07462-5 ]
In situ structure of the Nitrosopumilus maritimus S-layer - Composite map between C2 and C6
Resolution: 3.5 Å
EM Method: Subtomogram averaging
Fitted PDBs: 8c8r
Q-score: 0.438
von Kugelgen A, Cassidy CK, van Dorst S, Pagani LL, Batters C, Ford Z, Lowe J, Alva V, Stansfeld PJ, Bharat TAM
Nature (2024) 630 pp. 230-236 [ Pubmed: 38811725 DOI: doi:10.1038/s41586-024-07462-5 ]