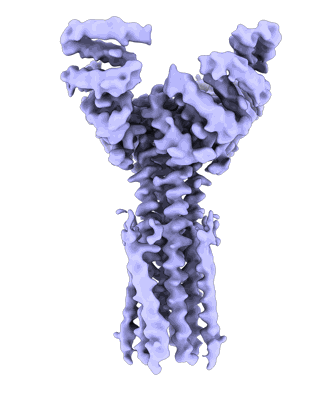
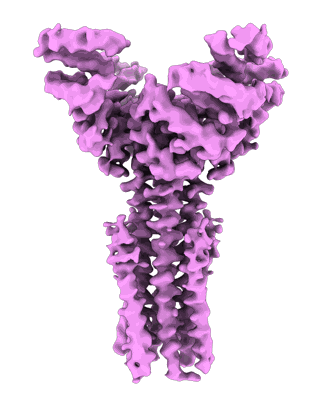
Artificial membrane protein TMHC4_R (ROCKET)
Abramsson M, Corey RA, Skerle J, Persson L, Anden O, Oluwole AO, Howard RJ, Lindahl E, Robinson CV, Strisovsky K, Marklund EG, Drew D, Stansfeld PJ, Landreh M
To Be Published
Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex
Nygaard R, Graham CLB, Belcher Dufrisne M, Colburn JD, Pepe J, Hydorn MA, Corradi S, Brown CM, Ashraf KU, Vickery ON, Briggs NS, Deering JJ, Kloss B, Botta B, Clarke OB, Columbus L, Dworkin J, Stansfeld PJ, Roper DI, Mancia F
Nat Commun (2023) 14 [ Pubmed: 37620344 DOI: 10.1038/s41467-023-40483-8 ]
Image sets:
Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8tj3
Q-score: 0.25
Nygaard R, Graham CLB, Belcher Dufrisne M, Colburn JD, Pepe J, Hydorn MA, Corradi S, Brown CM, Ashraf KU, Vickery ON, Briggs NS, Deering JJ, Kloss B, Botta B, Clarke OB, Columbus L, Dworkin J, Stansfeld PJ, Roper DI, Mancia F
Nat Commun (2023) 14 pp. 5151-5151 [ DOI: doi:10.1038/s41467-023-40483-8 Pubmed: 37620344 ]
- Peptidoglycan d,d-transpeptidase mrda (70 kDa, Protein from Escherichia coli)
- Roda-pbp2 (111 kDa, Complex from Escherichia coli)
- Peptidoglycan glycosyltransferase mrdb (40 kDa, Protein from Escherichia coli)
Nygaard R, Graham CLB, Belcher Dufrisne M, Colburn JD, Pepe J, Hydorn MA, Corradi S, Brown CM, Ashraf KU, Vickery ON, Briggs NS, Deering JJ, Kloss B, Botta B, Clarke OB, Columbus L, Dworkin J, Stansfeld PJ, Roper DI, Mancia F
Nat Commun (2023) 14 pp. 5151-5151 [ DOI: doi:10.1038/s41467-023-40483-8 Pubmed: 37620344 ]
- Roda-pbp2 (111 kDa, Complex from Escherichia coli)
Nygaard R, Graham CLB, Belcher Dufrisne M, Colburn JD, Pepe J, Hydorn MA, Corradi S, Brown CM, Ashraf KU, Vickery ON, Briggs NS, Deering JJ, Kloss B, Botta B, Clarke OB, Columbus L, Dworkin J, Stansfeld PJ, Roper DI, Mancia F
Nat Commun (2023) 14 pp. 5151-5151 [ DOI: doi:10.1038/s41467-023-40483-8 Pubmed: 37620344 ]
- Roda-pbp2 (111 kDa, Complex from Escherichia coli)
Single particle cryo-EM of KdpFABC WT (KdpB-Ser162-P) under turnover condition
Silberberg JM, Stock C, Hielkema L, Corey RA, Rheinberger J, Wunnicke D, Dubach VRA, Stansfeld PJ, Hänelt I, Paulino C
eLife (2022) 11 [ DOI: 10.7554/elife.80988 Pubmed: 36255052 ]
Image sets:
- Unaligned movie frames of the KdpFABC WT (KdpB-Ser162-P) under turnover conditions in DDM (Dataset 3)
- Unaligned movie frames of the KdpFABC WT (KdpB-Ser162-P) under turnover conditions in DDM (Dataset 4)
- Unaligned movie frames of the KdpFABC WT (KdpB-Ser162-P) under turnover conditions in DDM (Dataset 5)
- Unaligned movie frames of the KdpFABC WT (KdpB-Ser162-P) under turnover conditions in DDM (Dataset 2)
- Unaligned movie frames of the KdpFABC WT (KdpB-Ser162-P) under turnover conditions in DDM (Dataset 1)
Single particle cryo-EM of KdpFAB(Ser162-P, D307N)C in apo condition
Silberberg JM, Stock C, Hielkema L, Corey RA, Rheinberger J, Wunnicke D, Dubach VRA, Stansfeld PJ, Hänelt I, Paulino C
eLife (2022) 11 [ DOI: 10.7554/elife.80988 Pubmed: 36255052 ]
Image sets:
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 8)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 4)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 6)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 15)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 2)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 13)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 3)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 5)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 1)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 11)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 7)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 9)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 14)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 12)
- Unaligned movie frames of the KdpFAB(Ser162-P, D307N)C without nucleotide in DDM (Dataset 10)
Single particle cryo-EM of KdpFABC WT (KdpB-Ser162-P) in presence of orthovanadate
Silberberg JM, Stock C, Hielkema L, Corey RA, Rheinberger J, Wunnicke D, Dubach VRA, Stansfeld PJ, Hänelt I, Paulino C
eLife (2022) 11 [ DOI: 10.7554/elife.80988 Pubmed: 36255052 ]
Image sets:
Single particle cryo-EM of KdpFAB(S162A)C under turnover condition
Silberberg JM, Stock C, Hielkema L, Corey RA, Rheinberger J, Wunnicke D, Dubach VRA, Stansfeld PJ, Hänelt I, Paulino C
eLife (2022) 11 [ DOI: 10.7554/elife.80988 Pubmed: 36255052 ]
Image sets: