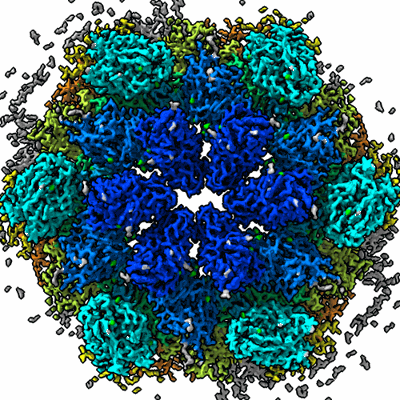
In situ structure of the Nitrosopumilus maritimus S-layer - Six-fold symmetry (C6)
Resolution: 3.4 Å
EM Method: Subtomogram averaging
Fitted PDBs: 8c8o
Q-score: 0.434
von Kugelgen A, Cassidy CK, van Dorst S, Pagani LL, Batters C, Ford Z, Lowe J, Alva V, Stansfeld PJ, Bharat TAM
Nature (2024) 630 pp. 230-236 [ Pubmed: 38811725 DOI: doi:10.1038/s41586-024-07462-5 ]
In situ structure of the Nitrosopumilus maritimus S-layer - Composite map between C2 and C6
Resolution: 3.5 Å
EM Method: Subtomogram averaging
Fitted PDBs: 8c8r
Q-score: 0.438
von Kugelgen A, Cassidy CK, van Dorst S, Pagani LL, Batters C, Ford Z, Lowe J, Alva V, Stansfeld PJ, Bharat TAM
Nature (2024) 630 pp. 230-236 [ Pubmed: 38811725 DOI: doi:10.1038/s41586-024-07462-5 ]
In vitro structure of the Nitrosopumilus maritimus S-layer - Six-fold symmetry (C6)
Resolution: 2.87 Å
EM Method: Single-particle
Fitted PDBs: 8c8k
Q-score: 0.543
von Kugelgen A, Cassidy CK, van Dorst S, Pagani LL, Batters C, Ford Z, Lowe J, Alva V, Stansfeld PJ, Bharat TAM
Nature (2024) 630 pp. 230-236 [ Pubmed: 38811725 DOI: doi:10.1038/s41586-024-07462-5 ]
In vitro structure of the Nitrosopumilus maritimus S-layer - Two-fold symmetry (C2)
Resolution: 2.71 Å
EM Method: Single-particle
Fitted PDBs: 8c8l
Q-score: 0.595
von Kugelgen A, Cassidy CK, van Dorst S, Pagani LL, Batters C, Ford Z, Lowe J, Alva V, Stansfeld PJ, Bharat TAM
Nature (2024) 630 pp. 230-236 [ Pubmed: 38811725 DOI: doi:10.1038/s41586-024-07462-5 ]
Resolution: 2.87 Å
EM Method: Single-particle
Fitted PDBs: 8c8m
Q-score: 0.556
von Kugelgen A, Cassidy CK, van Dorst S, Pagani LL, Batters C, Ford Z, Lowe J, Alva V, Stansfeld PJ, Bharat TAM
Nature (2024) 630 pp. 230-236 [ Pubmed: 38811725 DOI: doi:10.1038/s41586-024-07462-5 ]
In situ structure of the Nitrosopumilus maritimus S-layer - Two-fold symmetry (C2)
Resolution: 3.4 Å
EM Method: Subtomogram averaging
Fitted PDBs: 8c8n
Q-score: 0.489
von Kugelgen A, Cassidy CK, van Dorst S, Pagani LL, Batters C, Ford Z, Lowe J, Alva V, Stansfeld PJ, Bharat TAM
Nature (2024) 630 pp. 230-236 [ Pubmed: 38811725 DOI: doi:10.1038/s41586-024-07462-5 ]
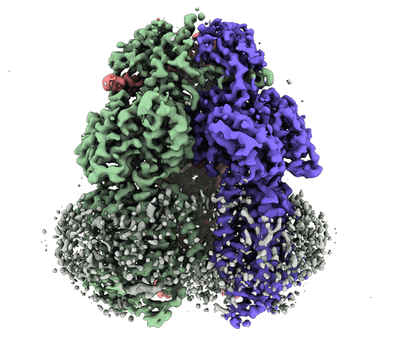
Structure of the Native Chemotaxis Core Signalling Complex from E-gene lysed E. coli cells.
Cassidy CK, Qin Z, Frosio T, Gosink K, Yang Z, Sansom MSP, Stansfeld PJ, Parkinson JS, Zhang P
Mbio (2023) 14 pp. e0079323-e0079323 [ DOI: doi:10.1128/mbio.00793-23 Pubmed: 37772839 ]
Cassidy CK, Qin Z, Frosio T, Gosink K, Yang Z, Sansom MSP, Stansfeld PJ, Parkinson JS, Zhang P
Mbio (2023) 14 pp. e0079323-e0079323 [ DOI: doi:10.1128/mbio.00793-23 Pubmed: 37772839 ]
Native Chemotaxis Core Signalling Complex from E. coli, Focused alignment on ligand binding domain
Cassidy CK, Qin Z, Frosio T, Gosink K, Yang Z, Sansom MSP, Stansfeld PJ, Parkinson JS, Zhang P
Mbio (2023) 14 pp. e0079323-e0079323 [ DOI: doi:10.1128/mbio.00793-23 Pubmed: 37772839 ]
Cryo-electron microscopy structure of the heavy metal efflux pump CusA in the symmetric closed state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7kf5
Q-score: 0.424
Moseng MA, Lyu M, Pipatpolkai T, Glaza P, Emerson CC, Stewart PL, Stansfeld PJ, Yu EW
Mbio (2021) 12 [ DOI: doi:10.1128/mBio.00452-21 Pubmed: 33820823 ]