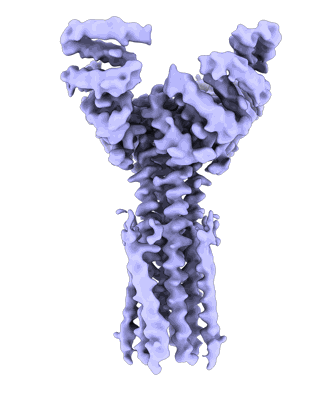
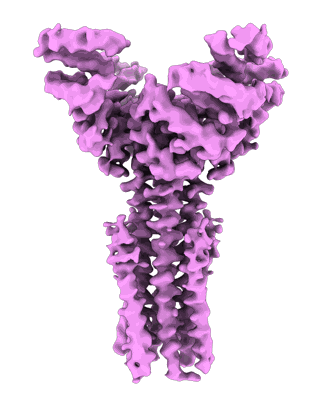
Artificial membrane protein TMHC4_R (ROCKET)
Abramsson M, Corey RA, Skerle J, Persson L, Anden O, Oluwole AO, Howard RJ, Lindahl E, Robinson CV, Strisovsky K, Marklund EG, Drew D, Stansfeld PJ, Landreh M
To Be Published
Structure of the Native Chemotaxis Core Signalling Complex from E-gene lysed E. coli cells.
Cassidy CK, Qin Z, Frosio T, Gosink K, Yang Z, Sansom MSP, Stansfeld PJ, Parkinson JS, Zhang P
Mbio (2023) 14 pp. e0079323-e0079323 [ Pubmed: 37772839 DOI: doi:10.1128/mbio.00793-23 ]
Cassidy CK, Qin Z, Frosio T, Gosink K, Yang Z, Sansom MSP, Stansfeld PJ, Parkinson JS, Zhang P
Mbio (2023) 14 pp. e0079323-e0079323 [ Pubmed: 37772839 DOI: doi:10.1128/mbio.00793-23 ]
Native Chemotaxis Core Signalling Complex from E. coli, Focused alignment on ligand binding domain
Cassidy CK, Qin Z, Frosio T, Gosink K, Yang Z, Sansom MSP, Stansfeld PJ, Parkinson JS, Zhang P
Mbio (2023) 14 pp. e0079323-e0079323 [ Pubmed: 37772839 DOI: doi:10.1128/mbio.00793-23 ]
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state
Resolution: 3.23 Å
EM Method: Single-particle
Fitted PDBs: 7tpg
Q-score: 0.497
Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper DI, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F
Nature (2022) 604 pp. 371-376 [ DOI: doi:10.1038/s41586-022-04555-x Pubmed: 35388216 ]
- Geranyl diphosphate (314 Da, Ligand)
- Ligand bound cmwaal with fab fragment bound (Complex)
- Fab light (l) chain (23 kDa, Protein from Homo sapiens)
- Fab heavy (h) chain (25 kDa, Protein from Homo sapiens)
- Putative cell surface polysaccharide polymerase/ligase (44 kDa, Protein from Cupriavidus metallidurans)
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state
Resolution: 3.46 Å
EM Method: Single-particle
Fitted PDBs: 7tpj
Q-score: 0.431
Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper DI, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F
Nature (2022) 604 pp. 371-376 [ DOI: doi:10.1038/s41586-022-04555-x Pubmed: 35388216 ]
- Fab heavy (h) chain (25 kDa, Protein from Homo sapiens)
- Putative cell surface polysaccharide polymerase/ligase (44 kDa, Protein from Cupriavidus metallidurans)
- Fab light (l) chain (23 kDa, Protein from Homo sapiens)
- Apo cmwaal with fab fragment bound (Complex)
Resolution: 4.37 Å
EM Method: Single-particle
Fitted PDBs: 7peo
Q-score: 0.298
Herdman M, von Kugelgen A, Kureisaite-Ciziene D, Duman R, El Omari K, Garman EF, Kjaer A, Kolokouris D, Lowe J, Wagner A, Stansfeld PJ, Bharat TAM
Structure (2022) 30 pp. 215- [ DOI: doi:10.1016/j.str.2021.10.012 Pubmed: 34800371 ]
- S-layer protein (25 kDa, Protein from Caulobacter vibrioides)
- Calcium ion (40 Da, Ligand)
- Holmium atom (164 Da, Ligand)
- Structure of the caulobacter crescentus s-layer protein rsaa n-terminal domain bound to lps and soaked with holmium (Complex from Caulobacter vibrioides)
In Situ Core-Signalling Unit of E. coli Chemoreceptor Array
Burt A, Cassidy CK, Ames P, Bacia-Verloop M, Baulard M, Huard K, Luthey-Schulten Z, Desfosses A, Stansfeld PJ, Margolin W, Parkinson JS, Gutsche I
Nat Commun (2020) 11 pp. 743-743 [ Pubmed: 32029744 DOI: doi:10.1038/s41467-020-14350-9 ]