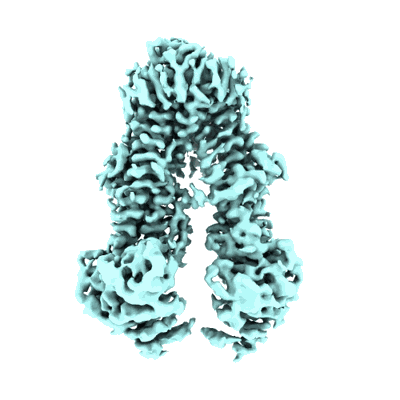
Structure of the wild-type Arabidopsis ABCB19 in the brassinolide-bound state
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8wom
Q-score: 0.451
Ying W, Wang Y, Wei H, Luo Y, Ma Q, Zhu H, Janssens H, Vukasinovic N, Kvasnica M, Winne JM, Gao Y, Tan S, Friml J, Liu X, Russinova E, Sun L
Science (2024) 383 pp. eadj4591-eadj4591 [ DOI: doi:10.1126/science.adj4591 Pubmed: 38513023 ]
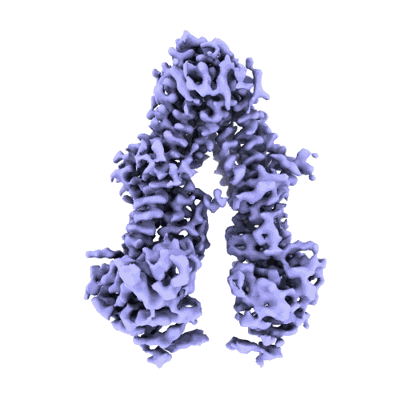
Structure of the wild-type Arabidopsis ABCB19 in the brassinolide and AMP-PNP bound state
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 8woo
Q-score: 0.401
Ying W, Wang Y, Wei H, Luo Y, Ma Q, Zhu H, Janssens H, Vukasinovic N, Kvasnica M, Winne JM, Gao Y, Tan S, Friml J, Liu X, Russinova E, Sun L
Science (2024) 383 pp. eadj4591-eadj4591 [ DOI: doi:10.1126/science.adj4591 Pubmed: 38513023 ]
- Abc transporter b family member 19 (136 kDa, Protein from Arabidopsis thaliana)
- Purified wild-type abcb19 protein in complex with brassinolide and amp-pnp (Cellular component from Arabidopsis thaliana)
- Magnesium ion (24 Da, Ligand)
- Brassinolide (480 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
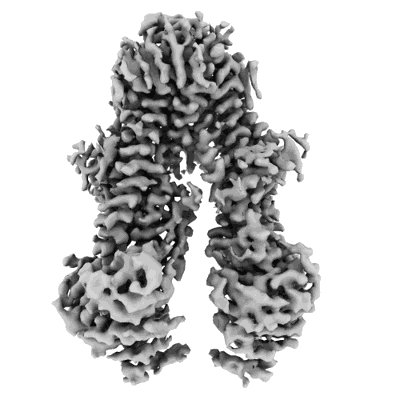
Structure of the Arabidopsis E529Q/E1174Q ABCB19 in the ATP bound state
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8wp0
Q-score: 0.464
Ying W, Wang Y, Wei H, Luo Y, Ma Q, Zhu H, Janssens H, Vukasinovic N, Kvasnica M, Winne JM, Gao Y, Tan S, Friml J, Liu X, Russinova E, Sun L
Science (2024) 383 pp. eadj4591-eadj4591 [ DOI: doi:10.1126/science.adj4591 Pubmed: 38513023 ]
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Abc transporter b family member 19 (136 kDa, Protein from Arabidopsis thaliana)
- Purified e529q/e1174q mutant abcb19 protein in complex with atp (Cellular component from Arabidopsis thaliana)
- Magnesium ion (24 Da, Ligand)
Structure of R2 with 3'UTR and DNA in binding state
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8ibw
Q-score: 0.385
Deng P, Tan SQ, Yang QY, Fu L, Wu Y, Zhu HZ, Sun L, Bao Z, Lin Y, Zhang QC, Wang H, Wang J, Liu JG
Cell (2023) 186 pp. 2865-2879.e20 [ DOI: doi:10.1016/j.cell.2023.05.032 Pubmed: 37301196 ]
- Zinc ion (65 Da, Ligand)
- Reverse transcriptase-like protein (123 kDa, Protein from Bombyx mori)
- 3'utr (16 kDa, RNA from Bombyx mori)
- Dna (60-mer) (18 kDa, DNA from Bombyx mori)
- R2 complex (177 kDa, Complex from Bombyx mori)
- Dna (60-mer) (18 kDa, DNA from Bombyx mori)
Structure of R2 with 3'UTR and DNA in unwinding state
Resolution: 3.74 Å
EM Method: Single-particle
Fitted PDBs: 8ibx
Q-score: 0.385
Deng P, Tan SQ, Yang QY, Fu L, Wu Y, Zhu HZ, Sun L, Bao Z, Lin Y, Zhang QC, Wang H, Wang J, Liu JG
Cell (2023) 186 pp. 2865-2879.e20 [ DOI: doi:10.1016/j.cell.2023.05.032 Pubmed: 37301196 ]
- Zinc ion (65 Da, Ligand)
- Reverse transcriptase-like protein (123 kDa, Protein from Bombyx mori)
- 3'utr (16 kDa, RNA from Bombyx mori)
- Dna (60-mer) (18 kDa, DNA from Bombyx mori)
- R2 complex (177 kDa, Complex from Bombyx mori)
- Dna (60-mer) (18 kDa, DNA from Bombyx mori)
Resolution: 3.47 Å
EM Method: Single-particle
Fitted PDBs: 8iby
Q-score: 0.342
Deng P, Tan SQ, Yang QY, Fu L, Wu Y, Zhu HZ, Sun L, Bao Z, Lin Y, Zhang QC, Wang H, Wang J, Liu JG
Cell (2023) 186 pp. 2865-2879.e20 [ DOI: doi:10.1016/j.cell.2023.05.032 Pubmed: 37301196 ]
- 5'orf rna (72 kDa, RNA from Bombyx mori)
- Zinc ion (65 Da, Ligand)
- Reverse transcriptase-like protein (123 kDa, Protein from Bombyx mori)
- R2 complex (177 kDa, Complex from Bombyx mori)
Structure of R2 with 5'ORF and 3'UTR
Resolution: 3.04 Å
EM Method: Single-particle
Fitted PDBs: 8ibz
Q-score: 0.435
Deng P, Tan SQ, Yang QY, Fu L, Wu Y, Zhu HZ, Sun L, Bao Z, Lin Y, Zhang QC, Wang H, Wang J, Liu JG
Cell (2023) 186 pp. 2865-2879.e20 [ DOI: doi:10.1016/j.cell.2023.05.032 Pubmed: 37301196 ]
- 5orf-linker-3utr (65 kDa, RNA from Bombyx mori)
- Zinc ion (65 Da, Ligand)
- Reverse transcriptase-like protein (123 kDa, Protein from Bombyx mori)
- R2 complex (177 kDa, Complex from Bombyx mori)
Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the NPA-bound state
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7y9u
Q-score: 0.492
Yang Z, Xia J, Hong J, Zhang C, Wei H, Ying W, Sun C, Sun L, Mao Y, Gao Y, Tan S, Friml J, Li D, Liu X, Sun L
Nature (2022) 609 pp. 611-615 [ DOI: doi:10.1038/s41586-022-05143-9 Pubmed: 35917925 ]
- 2-(naphthalen-1-ylcarbamoyl)benzoic acid (291 Da, Ligand)
- Atpin1 (Complex from Escherichia coli)
- Atpin1 in complex with a nanobody and npa (Complex from Arabidopsis thaliana)
- Nanobody (13 kDa, Protein from Escherichia coli)
- Nanobody (Complex)
- Auxin efflux carrier component 1 (67 kDa, Protein from Arabidopsis thaliana)
Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the IAA-bound state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7y9v
Q-score: 0.499
Yang Z, Xia J, Hong J, Zhang C, Wei H, Ying W, Sun C, Sun L, Mao Y, Gao Y, Tan S, Friml J, Li D, Liu X, Sun L
Nature (2022) 609 pp. 611-615 [ DOI: doi:10.1038/s41586-022-05143-9 Pubmed: 35917925 ]
- Water (18 Da, Ligand)
- Atpin1 (Complex from Escherichia coli)
- Atpin1 in complex with a nanobody and npa (Complex from Arabidopsis thaliana)
- Nanobody (13 kDa, Protein from Escherichia coli)
- Nanobody (Complex)
- 1h-indol-3-ylacetic acid (175 Da, Ligand)
- Auxin efflux carrier component 1 (67 kDa, Protein from Arabidopsis thaliana)