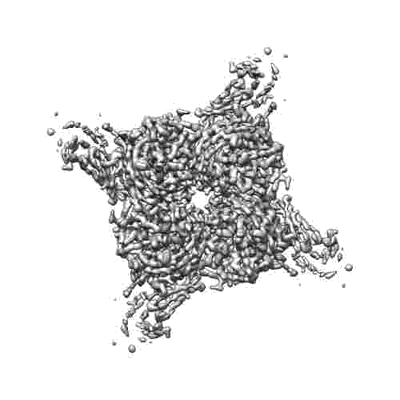
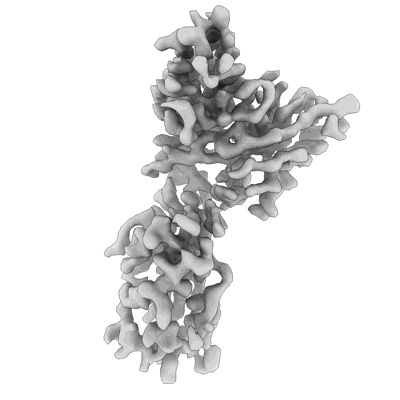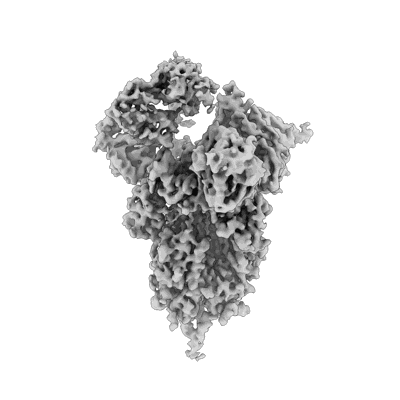N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 4 FNI19 Fab molecules
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8g30
Q-score: 0.487
Momont C, Dang HV, Zatta F, Hauser K, Wang C, di Iulio J, Minola A, Czudnochowski N, De Marco A, Branch K, Donermeyer D, Vyas S, Chen A, Ferri E, Guarino B, Powell AE, Spreafico R, Yim SS, Balce DR, Bartha I, Meury M, Croll TI, Belnap DM, Schmid MA, Schaiff WT, Miller JL, Cameroni E, Telenti A, Virgin HW, Rosen LE, Purcell LA, Lanzavecchia A, Snell G, Corti D, Pizzuto MS
Nature (2023) 618 pp. 590-597 [ Pubmed: 37258672 DOI: doi:10.1038/s41586-023-06136-y ]
- Calcium ion (40 Da, Ligand)
- N2 neuraminidase of a/tanzania/205/2010 h3n2 in complex with 4 fni19 fab molecules (420 kDa, Complex from Influenza A virus)
- Neuraminidase (54 kDa, Protein from Influenza A virus)
- Fni19 fab light chain (23 kDa, Protein from Homo sapiens)
- Fni19 fab heavy chain (24 kDa, Protein from Homo sapiens)
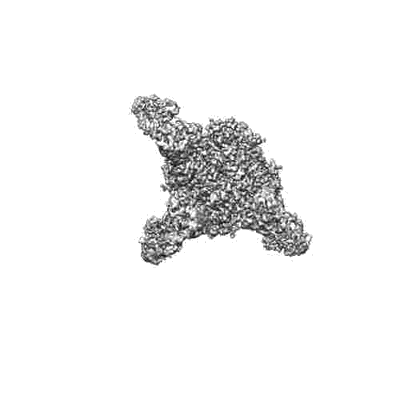
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 3 FNI9 Fab molecules
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8g3m
Q-score: 0.502
Momont C, Dang HV, Zatta F, Hauser K, Wang C, di Iulio J, Minola A, Czudnochowski N, De Marco A, Branch K, Donermeyer D, Vyas S, Chen A, Ferri E, Guarino B, Powell AE, Spreafico R, Yim SS, Balce DR, Bartha I, Meury M, Croll TI, Belnap DM, Schmid MA, Schaiff WT, Miller JL, Cameroni E, Telenti A, Virgin HW, Rosen LE, Purcell LA, Lanzavecchia A, Snell G, Corti D, Pizzuto MS
Nature (2023) 618 pp. 590-597 [ Pubmed: 37258672 DOI: doi:10.1038/s41586-023-06136-y ]
- Calcium ion (40 Da, Ligand)
- N2 neuraminidase of a/tanzania/205/2010 h3n2 in complex with 3 fni9 fab molecules (420 kDa, Complex from Influenza A virus)
- Fni9 fab light chain (23 kDa, Protein from Homo sapiens)
- Neuraminidase (54 kDa, Protein from Influenza A virus)
- Fni9 fab heavy chain (24 kDa, Protein from Homo sapiens)
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI9 Fab molecules
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8g3o
Q-score: 0.517
Momont C, Dang HV, Zatta F, Hauser K, Wang C, di Iulio J, Minola A, Czudnochowski N, De Marco A, Branch K, Donermeyer D, Vyas S, Chen A, Ferri E, Guarino B, Powell AE, Spreafico R, Yim SS, Balce DR, Bartha I, Meury M, Croll TI, Belnap DM, Schmid MA, Schaiff WT, Miller JL, Cameroni E, Telenti A, Virgin HW, Rosen LE, Purcell LA, Lanzavecchia A, Snell G, Corti D, Pizzuto MS
Nature (2023) 618 pp. 590-597 [ Pubmed: 37258672 DOI: doi:10.1038/s41586-023-06136-y ]
- Calcium ion (40 Da, Ligand)
- Neuraminidase (54 kDa, Protein from Influenza A virus)
- Fni9 fab light chain (23 kDa, Protein from Homo sapiens)
- 3 fni9 fabs in complex with tetrameric n2 na protein from a/hong_kong/2671/2019 strain (420 kDa, Complex from Influenza A virus)
- Fni9 fab heavy chain (24 kDa, Protein from Homo sapiens)
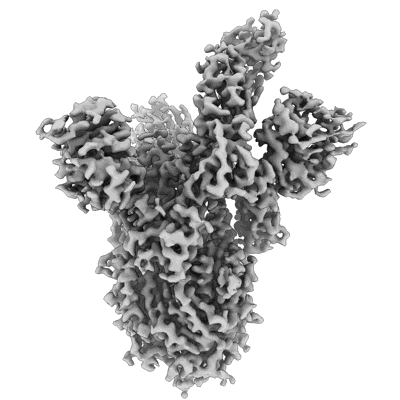
SARS-CoV-2 S glycoprotein in complex with S2X259 Fab
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7ra8
Q-score: 0.423
Tortorici MA, Czudnochowski N, Starr TN, Marzi R, Walls AC, Zatta F, Bowen JE, Jaconi S, Di Iulio J, Wang Z, De Marco A, Zepeda SK, Pinto D, Liu Z, Beltramello M, Bartha I, Housley MP, Lempp FA, Rosen LE, Dellota Jr E, Kaiser H, Montiel-Ruiz M, Zhou J, Addetia A, Guarino B, Culap K, Sprugasci N, Saliba C, Vetti E, Giacchetto-Sasselli I, Fregni CS, Abdelnabi R, Foo SC, Havenar-Daughton C, Schmid MA, Benigni F, Cameroni E, Neyts J, Telenti A, Virgin HW, Whelan SPJ, Snell G, Bloom JD, Corti D, Veesler D, Pizzuto MS
Nature (2021) 597 pp. 103-108 [ Pubmed: 34280951 DOI: doi:10.1038/s41586-021-03817-4 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 s glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Light chain fab s2x259 fab variable domain (11 kDa, Protein from Homo sapiens)
- S2x259 fab (Complex from Homo sapiens)
- Severe acute respiratory syndrome coronavirus 2 (Complex)
- Heavy chain fab s2x259 fab variable domain (13 kDa, Protein from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Resolution: 3.95 Å
EM Method: Single-particle
Fitted PDBs: 7r7n
Q-score: 0.411
Starr TN, Czudnochowski N, Liu Z, Zatta F, Park YJ, Addetia A, Pinto D, Beltramello M, Hernandez P, Greaney AJ, Marzi R, Glass WG, Zhang I, Dingens AS, Bowen JE, Tortorici MA, Walls AC, Wojcechowskyj JA, De Marco A, Rosen LE, Zhou J, Montiel-Ruiz M, Kaiser H, Dillen JR, Tucker H, Bassi J, Silacci-Fregni C, Housley MP, di Iulio J, Lombardo G, Agostini M, Sprugasci N, Culap K, Jaconi S, Meury M, Dellota Jr E, Abdelnabi R, Foo SC, Cameroni E, Stumpf S, Croll TI, Nix JC, Havenar-Daughton C, Piccoli L, Benigni F, Neyts J, Telenti A, Lempp FA, Pizzuto MS, Chodera JD, Hebner CM, Virgin HW, Whelan SPJ, Veesler D, Corti D, Bloom JD, Snell G
Nature (2021) 597 pp. 97-102 [ DOI: doi:10.1038/s41586-021-03807-6 Pubmed: 34261126 ]
- S2d106 fab heavy chain (13 kDa, Protein from Homo sapiens)
- S2d106 fab light chain (11 kDa, Protein from Homo sapiens)
- S2d106 (Complex from Homo sapiens)
- Sars-cov-2 spike in complex with the s2d106 neutralizing antibody fab fragment (Complex)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 spike in complex with the S2D106 neutralizing antibody Fab fragment
Starr TN, Czudnochowski N, Liu Z, Zatta F, Park YJ, Addetia A, Pinto D, Beltramello M, Hernandez P, Greaney AJ, Marzi R, Glass WG, Zhang I, Dingens AS, Bowen JE, Tortorici MA, Walls AC, Wojcechowskyj JA, De Marco A, Rosen LE, Zhou J, Montiel-Ruiz M, Kaiser H, Dillen JR, Tucker H, Bassi J, Silacci-Fregni C, Housley MP, di Iulio J, Lombardo G, Agostini M, Sprugasci N, Culap K, Jaconi S, Meury M, Dellota Jr E, Abdelnabi R, Foo SC, Cameroni E, Stumpf S, Croll TI, Nix JC, Havenar-Daughton C, Piccoli L, Benigni F, Neyts J, Telenti A, Lempp FA, Pizzuto MS, Chodera JD, Hebner CM, Virgin HW, Whelan SPJ, Veesler D, Corti D, Bloom JD, Snell G
Nature (2021) 597 pp. 97-102 [ DOI: doi:10.1038/s41586-021-03807-6 Pubmed: 34261126 ]
Starr TN, Czudnochowski N, Liu Z, Zatta F, Park YJ, Addetia A, Pinto D, Beltramello M, Hernandez P, Greaney AJ, Marzi R, Glass WG, Zhang I, Dingens AS, Bowen JE, Tortorici MA, Walls AC, Wojcechowskyj JA, De Marco A, Rosen LE, Zhou J, Montiel-Ruiz M, Kaiser H, Dillen JR, Tucker H, Bassi J, Silacci-Fregni C, Housley MP, di Iulio J, Lombardo G, Agostini M, Sprugasci N, Culap K, Jaconi S, Meury M, Dellota Jr E, Abdelnabi R, Foo SC, Cameroni E, Stumpf S, Croll TI, Nix JC, Havenar-Daughton C, Piccoli L, Benigni F, Neyts J, Telenti A, Lempp FA, Pizzuto MS, Chodera JD, Hebner CM, Virgin HW, Whelan SPJ, Veesler D, Corti D, Bloom JD, Snell G
Nature (2021) 597 pp. 97-102 [ DOI: doi:10.1038/s41586-021-03807-6 Pubmed: 34261126 ]