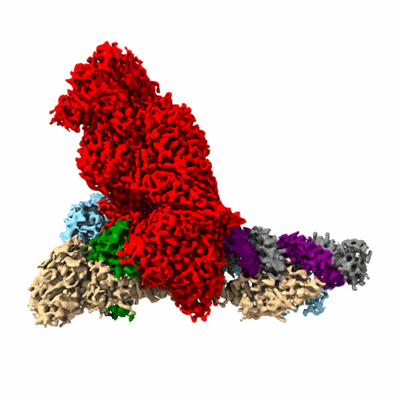
Cryo-EM structure of PEIP-Bs_enolase complex
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7xml
Q-score: 0.468
Zhang K, Li S, Wang Y, Wang Z, Mulvenna N, Yang H, Zhang P, Chen H, Li Y, Wang H, Gao Y, Wigneshweraraj S, Matthews S, Zhang K, Liu B
Cell Rep (2022) 40 pp. 111026-111026 [ DOI: doi:10.1016/j.celrep.2022.111026 Pubmed: 35793626 ]
- Peip-bs_enolase complex (100 kDa, Complex)
- Enolase (46 kDa, Protein from Bacillus subtilis (strain 168))
- Bs_enolase (Complex from Bacillus subtilis (strain 168))
- Putative gene 60 protein (8 kDa, Protein from Bacillus phage SP01)
- Magnesium ion (24 Da, Ligand)
- Peip (Complex from Bacillus phage SP01)
SARS-CoV-2 RdRP catalytic complex with T33-1 RNA
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7dte
Q-score: 0.528
Wu J, Wang H, Liu Q, Li R, Gao Y, Fang X, Zhong Y, Wang M, Wang Q, Rao Z, Gong P
Cell Rep (2021) 37 pp. 109882-109882 [ DOI: doi:10.1016/j.celrep.2021.109882 Pubmed: 34653416 ]
- Sars-cov-2 rna-dependent rna polymerase (Complex)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna-directed rna polymerase (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Rna (57-mer) (18 kDa, RNA from Foot-and-mouth disease virus)
- Rna (33-mer) (10 kDa, RNA from Foot-and-mouth disease virus)
- Sars-cov-2 rna-dependent rna polymerase catalytic complex (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rna (Complex)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM of peptide-like filament of 1-KMe3
Resolution: 4.3 Å
EM Method: Helical reconstruction
Fitted PDBs: 6x5i
Q-score: 0.34
Feng Z, Wang H, Wang F, Oh Y, Berciu C, Cui Q, Egelman EH, Xu B
Cell Rep Phys Sci (2020) 1 [ Pubmed: 32776017 DOI: doi:10.1016/j.xcrp.2020.100085 ]
- 1-kme3 peptide-like fibril (880 Da, Protein from Homo sapiens)
- 1-kme3 peptide-like fibril (Complex from Homo sapiens)
Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8
Resolution: 4.5 Å
EM Method: Single-particle
Fitted PDBs: 6ira
Q-score: 0.227
Zhang JB, Chang S, Xu P, Miao M, Wu H, Zhang Y, Zhang T, Wang H, Zhang J, Xie C, Song N, Luo C, Zhang X, Zhu S
Cell Rep (2018) 25 pp. 3582-3590.e4 [ DOI: doi:10.1016/j.celrep.2018.11.071 Pubmed: 30590034 ]
- Glutamate receptor ionotropic, nmda 2a (95 kDa, Protein from Homo sapiens)
- Glutamate receptor ionotropic, nmda 1 (95 kDa, Protein from Homo sapiens)
- Human glun1/glun2a nmda receptors in the glutamate/glycine-bound state at ph 7.8 (380 kDa, Complex from Homo sapiens)
Resolution: 5.1 Å
EM Method: Single-particle
Fitted PDBs: 6irf
Q-score: 0.158
Resolution: 5.5 Å
EM Method: Single-particle
Fitted PDBs: 6irg
Q-score: 0.147
Resolution: 7.8 Å
EM Method: Single-particle
Fitted PDBs: 6irh
Q-score: 0.171