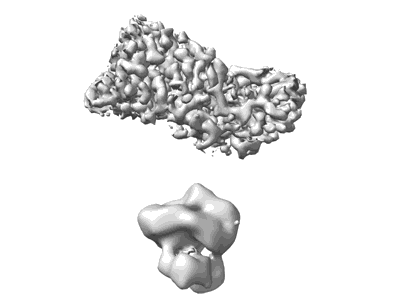
CryoEM density map of partial Rabies Virus nucleocapsid
Cai X, Zhou K, Alvarez-Cabrera AL, Si Z, Wang H, He Y, Li C, Zhou ZH
Viruses (2024) 16 [ DOI: doi:10.3390/v16091447 Pubmed: 39339924 ]
Resolution: 3.26 Å
EM Method: Helical reconstruction
Fitted PDBs: 8sxz
Q-score: 0.402
Jimah JR, Kundu N, Stanton AE, Sochacki KA, Canagarajah B, Chan L, Strub MP, Wang H, Taraska JW, Hinshaw JE
Dev Cell (2024) 59 pp. 1783- [ DOI: doi:10.1016/j.devcel.2024.04.008 Pubmed: 38663399 ]
- Gdp-bound human dynamin helical polymer assembled on the membrane (85 MDa, Complex from Homo sapiens)
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Dynamin-1 (86 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
Resolution: 2.86 Å
EM Method: Helical reconstruction
Fitted PDBs: 8sz4
Q-score: 0.344
Jimah JR, Kundu N, Stanton AE, Sochacki KA, Canagarajah B, Chan L, Strub MP, Wang H, Taraska JW, Hinshaw JE
Dev Cell (2024) 59 pp. 1783- [ DOI: doi:10.1016/j.devcel.2024.04.008 Pubmed: 38663399 ]
- Gdp-bound human dynamin helical polymer assembled on the membrane (85 MDa, Complex from Homo sapiens)
- Dynamin-1 (86 kDa, Protein from Homo sapiens)
Resolution: 2.84 Å
EM Method: Helical reconstruction
Fitted PDBs: 8sz7
Q-score: 0.308
Jimah JR, Kundu N, Stanton AE, Sochacki KA, Canagarajah B, Chan L, Strub MP, Wang H, Taraska JW, Hinshaw JE
Dev Cell (2024) 59 pp. 1783- [ Pubmed: 38663399 DOI: doi:10.1016/j.devcel.2024.04.008 ]
- Dynamin-1 (86 kDa, Protein from Homo sapiens)
- Gdp-bound human dynamin helical polymer assembled on the membrane (85 MDa, Complex from Homo sapiens)
Resolution: 3.55 Å
EM Method: Helical reconstruction
Fitted PDBs: 8sz8
Q-score: 0.358
Jimah JR, Kundu N, Stanton AE, Sochacki KA, Canagarajah B, Chan L, Strub MP, Wang H, Taraska JW, Hinshaw JE
Dev Cell (2024) 59 pp. 1783- [ DOI: doi:10.1016/j.devcel.2024.04.008 Pubmed: 38663399 ]
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Dynamin-1 (86 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Cryo-em of the gdp-bound human dynamin polymer assembled on the membrane in the super constricted state (full helix) (85 MDa, Complex from Homo sapiens)
Resolution: 3.58 Å
EM Method: Helical reconstruction
Fitted PDBs: 8t0k
Q-score: 0.368
Jimah JR, Kundu N, Stanton AE, Sochacki KA, Canagarajah B, Chan L, Strub MP, Wang H, Taraska JW, Hinshaw JE
Dev Cell (2024) 59 pp. 1783- [ DOI: doi:10.1016/j.devcel.2024.04.008 Pubmed: 38663399 ]
- Dynamin-1 (98 kDa, Protein from Homo sapiens)
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Gdp-bound human dynamin (full-length) helical polymer assembled on the membrane (98 MDa, Complex from Homo sapiens)
Resolution: 3.97 Å
EM Method: Helical reconstruction
Fitted PDBs: 8t0r
Q-score: 0.336
Jimah JR, Kundu N, Stanton AE, Sochacki KA, Canagarajah B, Chan L, Strub MP, Wang H, Taraska JW, Hinshaw JE
Dev Cell (2024) 59 pp. 1783- [ DOI: doi:10.1016/j.devcel.2024.04.008 Pubmed: 38663399 ]
- Dynamin-1 (98 kDa, Protein from Homo sapiens)
- Gdp-bound human dynamin (full-length) helical polymer assembled on the membrane (full helix) (98 MDa, Complex from Homo sapiens)
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
Cryo-EM structure of alpha-synuclein gS87 fibril
Resolution: 3.1 Å
EM Method: Helical reconstruction
Fitted PDBs: 8jex
Q-score: 0.537
Hu J, Xia W, Zeng S, Lim YJ, Tao Y, Sun Y, Zhao L, Wang H, Le W, Li D, Zhang S, Liu C, Li YM
Nat Commun (2024) 15 pp. 2677-2677 [ Pubmed: 38538591 DOI: doi:10.1038/s41467-024-46898-1 ]
- Alpha-synuclein (14 kDa, Protein from Homo sapiens)
- Unverified amino acid chain (783 Da, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cryo-em structure of alpha-synuclein gs87 fibril (Cellular component from Homo sapiens)
Cryo-EM structure of alpha-synuclein pS87 fibril
Resolution: 2.6 Å
EM Method: Helical reconstruction
Fitted PDBs: 8jey
Q-score: 0.587
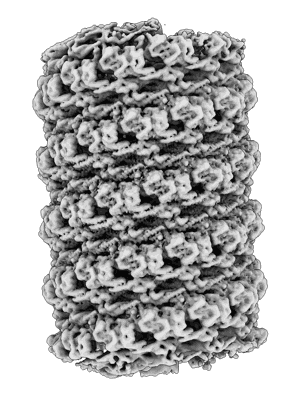
Membrane-bound CHMP2A-CHMP3 filament (430 Angstrom diameter)
Resolution: 3.3 Å
EM Method: Helical reconstruction
Fitted PDBs: 7zcg
Q-score: 0.454
Azad K, Guilligay D, Boscheron C, Maity S, De Franceschi N, Sulbaran G, Effantin G, Wang H, Kleman JP, Bassereau P, Schoehn G, Roos WH, Desfosses A, Weissenhorn W
Nat Struct Mol Biol (2023) 30 pp. 81-90 [ Pubmed: 36604498 DOI: doi:10.1038/s41594-022-00867-8 ]