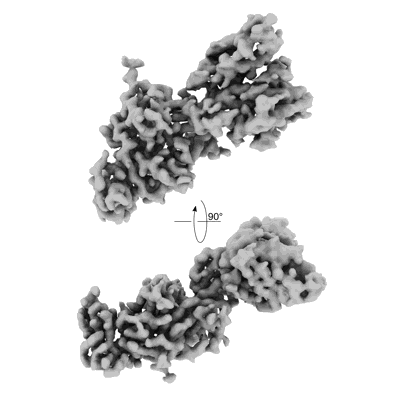Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 9asd
Q-score: 0.506
Rosen LE, Tortorici MA, De Marco A, Pinto D, Foreman WB, Taylor AL, Park YJ, Bohan D, Rietz T, Errico JM, Hauser K, Dang HV, Chartron JW, Giurdanella M, Cusumano G, Saliba C, Zatta F, Sprouse KR, Addetia A, Zepeda SK, Brown J, Lee J, Dellota Jr E, Rajesh A, Noack J, Tao Q, DaCosta Y, Tsu B, Acosta R, Subramanian S, de Melo GD, Kergoat L, Zhang I, Liu Z, Guarino B, Schmid MA, Schnell G, Miller JL, Lempp FA, Czudnochowski N, Cameroni E, Whelan SPJ, Bourhy H, Purcell LA, Benigni F, di Iulio J, Pizzuto MS, Lanzavecchia A, Telenti A, Snell G, Corti D, Veesler D, Starr TN
Cell (2024) 187 pp. 7196- [ Pubmed: 39383863 DOI: doi:10.1016/j.cell.2024.09.026 ]
- Spike glycoprotein (141 kDa, Protein from Homo sapiens)
- Vir-7229 fab light chain (11 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ba 2.86 rbd - s2v29 complex (local refinement) (Complex from Sarbecovirus)
- Vir-7229 fab heavy chain (13 kDa, Protein from Homo sapiens)
VIR-7229 Fab fragment bound the BA.2.86 spike trimer (global refinement)
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 9au2
Q-score: 0.499
Rosen LE, Tortorici MA, De Marco A, Pinto D, Foreman WB, Taylor AL, Park YJ, Bohan D, Rietz T, Errico JM, Hauser K, Dang HV, Chartron JW, Giurdanella M, Cusumano G, Saliba C, Zatta F, Sprouse KR, Addetia A, Zepeda SK, Brown J, Lee J, Dellota Jr E, Rajesh A, Noack J, Tao Q, DaCosta Y, Tsu B, Acosta R, Subramanian S, de Melo GD, Kergoat L, Zhang I, Liu Z, Guarino B, Schmid MA, Schnell G, Miller JL, Lempp FA, Czudnochowski N, Cameroni E, Whelan SPJ, Bourhy H, Purcell LA, Benigni F, di Iulio J, Pizzuto MS, Lanzavecchia A, Telenti A, Snell G, Corti D, Veesler D, Starr TN
Cell (2024) 187 pp. 7196- [ Pubmed: 39383863 DOI: doi:10.1016/j.cell.2024.09.026 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand from Severe acute respiratory syndrome coronavirus 2)
- Ba.2.86 spike glycoprotein in complex with vir-7229 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Vir-7229 fab heavy chain (24 kDa, Protein from Homo sapiens)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Vir-7229 fab light chain (23 kDa, Protein from Homo sapiens)
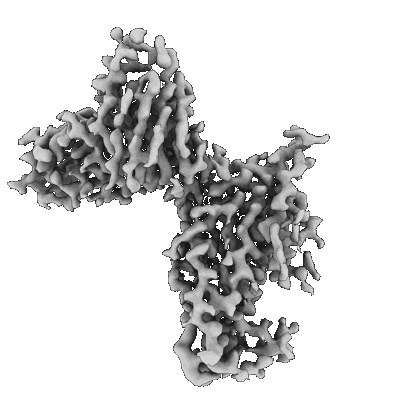
SARS-CoV-2 Spike NTD in complex with neutralizing Fab SARS2-57 (local refinement)
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 7sww
Q-score: 0.328
Adams LJ, VanBlargan LA, Liu Z, Gilchuk P, Zhao H, Chen RE, Raju S, Whitener B, Shrihari S, Jethva PN, Gross ML, Crowe JE, Whelan SPJ, Diamond MS, Fremont DH
To Be Published
- Spike protein s1 (33 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike bound by sars2-57 fab (Complex)
- Sars2-57 fv light chain (12 kDa, Protein from Mus musculus)
- Sars2-57 fv heavy chain (12 kDa, Protein from Mus musculus)
SARS-CoV-2 Spike in complex with neutralizing Fab SARS2-57 (three down conformation)
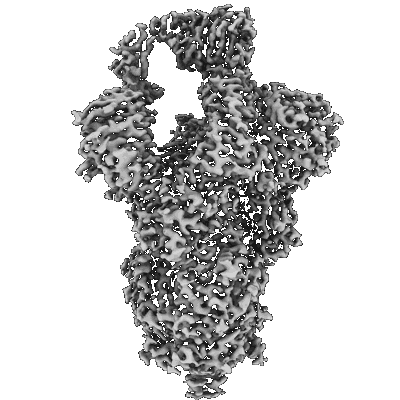
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 7swx
Q-score: 0.446
Adams LJ, VanBlargan LA, Liu Z, Gilchuk P, Zhao H, Chen RE, Raju S, Whitener B, Shrihari S, Jethva PN, Gross ML, Crowe JE, Whelan SPJ, Diamond MS, Fremont DH
To Be Published
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike bound by sars2-57 fab (Complex)
- Sars2-57 fv light chain (12 kDa, Protein from Mus musculus)
- Spike glycoprotein (125 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars2-57 fv heavy chain (12 kDa, Protein from Mus musculus)
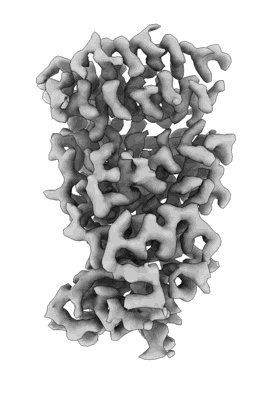
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8erq
Q-score: 0.493
Park YJ, Pinto D, Walls AC, Liu Z, De Marco A, Benigni F, Zatta F, Silacci-Fregni C, Bassi J, Sprouse KR, Addetia A, Bowen JE, Stewart C, Giurdanella M, Saliba C, Guarino B, Schmid MA, Franko NM, Logue JK, Dang HV, Hauser K, di Iulio J, Rivera W, Schnell G, Rajesh A, Zhou J, Farhat N, Kaiser H, Montiel-Ruiz M, Noack J, Lempp FA, Janer J, Abdelnabi R, Maes P, Ferrari P, Ceschi A, Giannini O, de Melo GD, Kergoat L, Bourhy H, Neyts J, Soriaga L, Purcell LA, Snell G, Whelan SPJ, Lanzavecchia A, Virgin HW, Piccoli L, Chu HY, Pizzuto MS, Corti D, Veesler D
Science (2022) 378 pp. 619-627 [ Pubmed: 36264829 DOI: doi:10.1126/science.adc9127 ]
- Sars-cov-2 ba.1 spike ectodomain trimer in complex with the s2x324 neutralizing antibody fab fragment (local refinement of the rbd and s2x324) (Complex from Severe acute respiratory syndrome coronavirus 2)
- S2x324 fab heavy chain (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 omicron ba.1 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- S2x324 fab light chain (11 kDa, Protein from Homo sapiens)
- S2x324 fab (Complex from Homo sapiens)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8err
Q-score: 0.449
Park YJ, Pinto D, Walls AC, Liu Z, De Marco A, Benigni F, Zatta F, Silacci-Fregni C, Bassi J, Sprouse KR, Addetia A, Bowen JE, Stewart C, Giurdanella M, Saliba C, Guarino B, Schmid MA, Franko NM, Logue JK, Dang HV, Hauser K, di Iulio J, Rivera W, Schnell G, Rajesh A, Zhou J, Farhat N, Kaiser H, Montiel-Ruiz M, Noack J, Lempp FA, Janer J, Abdelnabi R, Maes P, Ferrari P, Ceschi A, Giannini O, de Melo GD, Kergoat L, Bourhy H, Neyts J, Soriaga L, Purcell LA, Snell G, Whelan SPJ, Lanzavecchia A, Virgin HW, Piccoli L, Chu HY, Pizzuto MS, Corti D, Veesler D
Science (2022) 378 pp. 619-627 [ Pubmed: 36264829 DOI: doi:10.1126/science.adc9127 ]
- S2x324 light chain (11 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 omicron ba.1 spike ectodomain trimer in complex with the s2x324 neutralizing antibody fab fragment (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 omicron ba.1 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- S2x324 fab (Complex from Homo sapiens)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- S2x324 heavy chain (13 kDa, Protein from Homo sapiens)
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7uhb
Q-score: 0.366
Hunt AC, Case JB, Park YJ, Cao L, Wu K, Walls AC, Liu Z, Bowen JE, Yeh HW, Saini S, Helms L, Zhao YT, Hsiang TY, Starr TN, Goreshnik I, Kozodoy L, Carter L, Ravichandran R, Green LB, Matochko WL, Thomson CA, Vogeli B, Kruger A, VanBlargan LA, Chen RE, Ying B, Bailey AL, Kafai NM, Boyken SE, Ljubetic A, Edman N, Ueda G, Chow CM, Johnson M, Addetia A, Navarro MJ, Panpradist N, Gale Jr M, Freedman BS, Bloom JD, Ruohola-Baker H, Whelan SPJ, Stewart L, Diamond MS, Veesler D, Jewett MC, Baker D
Sci Transl Med (2022) 14 pp. eabn1252-eabn1252 [ Pubmed: 35412328 DOI: doi:10.1126/scitranslmed.abn1252 ]
- Sars-cov-2 spike in complex with ahb2-2gs-sb175 (local refinement of the rbd and ahb2) (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- Multivalent miniprotein inhibitor ahb2-2gs-sb175 (17 kDa, Protein from synthetic construct)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
- Ahb2-2gs-sb175 (Complex from Synthetic construct)
SARS-CoV-2 spike in complex with AHB2-2GS-SB175
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7uhc
Q-score: 0.465
Hunt AC, Case JB, Park YJ, Cao L, Wu K, Walls AC, Liu Z, Bowen JE, Yeh HW, Saini S, Helms L, Zhao YT, Hsiang TY, Starr TN, Goreshnik I, Kozodoy L, Carter L, Ravichandran R, Green LB, Matochko WL, Thomson CA, Vogeli B, Kruger A, VanBlargan LA, Chen RE, Ying B, Bailey AL, Kafai NM, Boyken SE, Ljubetic A, Edman N, Ueda G, Chow CM, Johnson M, Addetia A, Navarro MJ, Panpradist N, Gale Jr M, Freedman BS, Bloom JD, Ruohola-Baker H, Whelan SPJ, Stewart L, Diamond MS, Veesler D, Jewett MC, Baker D
Sci Transl Med (2022) 14 pp. eabn1252-eabn1252 [ Pubmed: 35412328 DOI: doi:10.1126/scitranslmed.abn1252 ]
- Multivalent miniprotein inhibitor ahb2-2gs-sb175 (Complex from Synthetic construct)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Multivalent miniprotein inhibitor ahb2-2gs-sb175 (17 kDa, Protein from synthetic construct)
- Sars-cov-2 spike in complex with multivalent minibinder ahb2-2gs-sb175 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 hexapro spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
Structure of vesicular stomatitis virus (helical reconstruction, 4.1 A resolution)
Resolution: 4.1 Å
EM Method: Helical reconstruction
Fitted PDBs: 7umk
Q-score: 0.311
Jenni S, Horwitz JA, Bloyet LM, Whelan SPJ, Harrison SC
Nat Commun (2022) 13 pp. 4802-4802 [ DOI: doi:10.1038/s41467-022-32223-1 Pubmed: 35970826 ]
- Matrix protein (26 kDa, Protein from Vesicular stomatitis Indiana virus)
- Rna (5'-r(p*up*up*up*up*up*up*up*up*u)-3') (2 kDa, RNA from Vesicular stomatitis Indiana virus)
- Nucleoprotein (47 kDa, Protein from Vesicular stomatitis Indiana virus)
- Vesicular stomatitis indiana virus (Virus from Vesicular stomatitis Indiana virus)
Structure of vesicular stomatitis virus (local reconstruction, 3.5 A resolution)
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7uml
Q-score: 0.399
Jenni S, Horwitz JA, Bloyet LM, Whelan SPJ, Harrison SC
Nat Commun (2022) 13 pp. 4802-4802 [ DOI: doi:10.1038/s41467-022-32223-1 Pubmed: 35970826 ]
- Matrix protein (26 kDa, Protein from Vesicular stomatitis Indiana virus)
- Nucleoprotein (47 kDa, Protein from Vesicular stomatitis Indiana virus)
- Vesicular stomatitis indiana virus (Virus from Vesicular stomatitis Indiana virus)
- Rna (5'-r(p*up*up*up*up*up*up*up*up*up*up*up*up*u)-3') (3 kDa, RNA from Vesicular stomatitis Indiana virus)