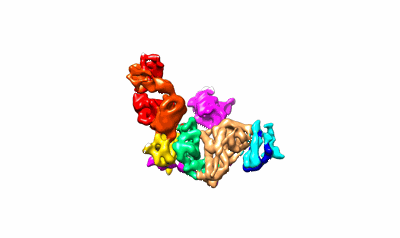
Prefusion RSV F Bound to Lonafarnib and D25 Fab
Resolution: 3.17 Å
EM Method: Single-particle
Fitted PDBs: 8kg5
Q-score: 0.499
Yang Q, Xue B, Liu F, Lu Y, Tang J, Yan M, Wu Q, Chen R, Zhou A, Liu L, Liu J, Qu C, Wu Q, Fu M, Zhong J, Dong J, Chen S, Wang F, Zhou Y, Zheng J, Peng W, Shang J, Chen X
Signal Transduct Target Ther (2024) 9 pp. 144-144 [ Pubmed: 38853183 DOI: doi:10.1038/s41392-024-01858-5 ]
- D25 light chain (23 kDa, Protein from Homo sapiens)
- Fusion glycoprotein f0,fibritin (64 kDa, Protein from Enterobacteria phage T4)
- 4-{2-[4-(3,10-dibromo-8-chloro-6,11-dihydro-5h-benzo[5,6]cyclohepta[1,2-b]pyridin-11-yl)piperidin-1-yl]-2-oxoethyl}piperidine-1-carboxamide (638 Da, Ligand)
- D25 heavy chain (24 kDa, Protein from Homo sapiens)
- Prefusion rsv f bound to lonafarnib and d25 fab (Complex from Human respiratory syncytial virus A (strain A2))
Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement)
Resolution: 3.64 Å
EM Method: Single-particle
Fitted PDBs: 7yoy
Q-score: 0.464
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ Pubmed: 37992686 DOI: doi:10.1016/j.xcrm.2023.101296 ]
- 5e3,3e8 (Complex from Oryctolagus cuniculus)
- 5e3 heavy chain (13 kDa, Protein from Oryctolagus cuniculus)
- Ebv ghgl-gp42 in complex with mabs 3e8 and 5e3 (Complex)
- Soluble gp42 (15 kDa, Protein from Human gammaherpesvirus 4)
- Gp42 (Complex from Human gammaherpesvirus 4)
- 3e8 heavy chain (12 kDa, Protein from Oryctolagus cuniculus)
- 5e3 light chain (11 kDa, Protein from Oryctolagus cuniculus)
- 3e8 light chain (11 kDa, Protein from Oryctolagus cuniculus)
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement)
Resolution: 3.54 Å
EM Method: Single-particle
Fitted PDBs: 7yp1
Q-score: 0.523
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ Pubmed: 37992686 DOI: doi:10.1016/j.xcrm.2023.101296 ]
- Gh,gl (Complex from Human gammaherpesvirus 4)
- 10e4 (Complex from Oryctolagus cuniculus)
- Ebv gl (8 kDa, Protein from Human gammaherpesvirus 4)
- 10e4 light chain (11 kDa, Protein from Oryctolagus cuniculus)
- Ebv gh (22 kDa, Protein from Human gammaherpesvirus 4)
- 10e4 heavy chain (12 kDa, Protein from Oryctolagus cuniculus)
- Ghgl-gp42 in complex with mab 10e4 (Complex)
Cryo-EM density map of EBV gHgL-gp42 in complex with four mAbs 5E3, 3E8, 6H2 and 10E4
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ Pubmed: 37992686 DOI: doi:10.1016/j.xcrm.2023.101296 ]
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement)
Resolution: 3.52 Å
EM Method: Single-particle
Fitted PDBs: 7yp2
Q-score: 0.53
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ Pubmed: 37992686 DOI: doi:10.1016/j.xcrm.2023.101296 ]
- Gh (Complex from Human gammaherpesvirus 4)
- 6h2 (Complex from Mus musculus)
- Envelope glycoprotein h (34 kDa, Protein from Human gammaherpesvirus 4)
- 6h2 light chain (12 kDa, Protein from Mus musculus)
- Ebv ghgl-gp42 in complex with mab 6h2 (Complex)
- 6h2 heavy chain (13 kDa, Protein from Mus musculus)
Cryo-EM structure of CopC-CaM-caspase-3 with NAD+
Resolution: 3.35 Å
EM Method: Single-particle
Fitted PDBs: 7xn4
Q-score: 0.492
Zhang K, Peng T, Tao X, Tian M, Li Y, Wang Z, Ma S, Hu S, Pan X, Xue J, Luo J, Wu Q, Fu Y, Li S
Mol Cell (2022) 82 pp. 4712-4726.e7 [ Pubmed: 36423631 DOI: doi:10.1016/j.molcel.2022.10.032 ]
- Cryo-em structure of copc-cam-caspase-3 with nad+ (Complex from Homo sapiens)
- Calmodulin-1 (16 kDa, Protein from Homo sapiens)
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Arginine adp-riboxanase copc (52 kDa, Protein from Chromobacterium violaceum)
- Caspase-3 (31 kDa, Protein from Homo sapiens)
Cryo-EM structure of CopC-CaM-caspase-3 with ADPR
Resolution: 3.18 Å
EM Method: Single-particle
Fitted PDBs: 7xn5
Q-score: 0.517
Zhang K, Peng T, Tao X, Tian M, Li Y, Wang Z, Ma S, Hu S, Pan X, Xue J, Luo J, Wu Q, Fu Y, Li S
Mol Cell (2022) 82 pp. 4712-4726.e7 [ Pubmed: 36423631 DOI: doi:10.1016/j.molcel.2022.10.032 ]
- Nicotinamide (122 Da, Ligand)
- Calmodulin-1 (16 kDa, Protein from Homo sapiens)
- Cryo-em structure of copc-cam-caspase-3 with adpr (Complex from Homo sapiens)
- Arginine adp-riboxanase copc (52 kDa, Protein from Chromobacterium violaceum)
- Caspase-3 (31 kDa, Protein from Homo sapiens)
- Adenosine-5-diphosphoribose (559 Da, Ligand)
Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization
Resolution: 3.45 Å
EM Method: Single-particle
Fitted PDBs: 7xn6
Q-score: 0.466
Zhang K, Peng T, Tao X, Tian M, Li Y, Wang Z, Ma S, Hu S, Pan X, Xue J, Luo J, Wu Q, Fu Y, Li S
Mol Cell (2022) 82 pp. 4712-4726.e7 [ Pubmed: 36423631 DOI: doi:10.1016/j.molcel.2022.10.032 ]
- Nicotinamide (122 Da, Ligand)
- Calmodulin-1 (16 kDa, Protein from Homo sapiens)
- Caspase-3 (32 kDa, Protein from Homo sapiens)
- Arginine adp-riboxanase copc (52 kDa, Protein from Chromobacterium violaceum)
- Cryo-em structure of copc-cam-caspase-3 with adpr-deacylization (Complex from Homo sapiens)
Cryo-EM structure of EBV glycoprotein complex gHgL-gp42 bound by a neutralizing antibody 6H2
Cryo-EM maps of the (TGA3)2-(NPR1)2-(TGA3)2 complex
Kumar S, Zavaliev R, Wu Q, Zhou Y, Cheng J, Dillard L, Powers J, Withers J, Zhao J, Guan Z, Borgnia MJ, Bartesaghi A, Dong X, Zhou P
Nature (2022) 605 pp. 561-566 [ Pubmed: 35545668 DOI: doi:10.1038/s41586-022-04699-w ]