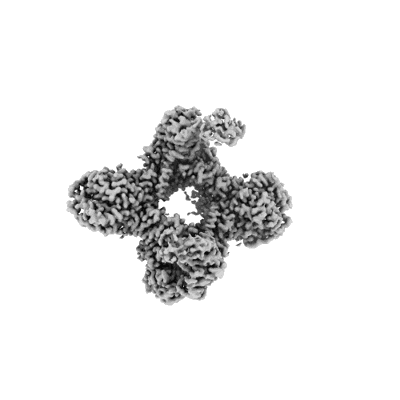
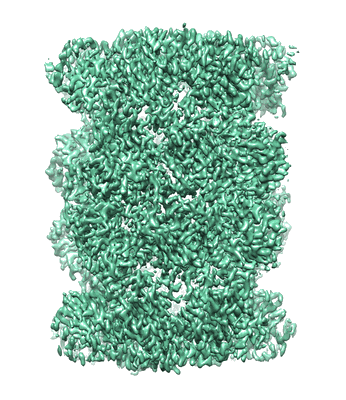Structure of Gabija AB complex
Resolution: 2.79 Å
EM Method: Single-particle
Fitted PDBs: 8tjy
Q-score: 0.534
Yang XY, Shen Z, Xie J, Greenwald J, Marathe I, Lin Q, Xie WJ, Wysocki VH, Fu TM
Nat Struct Mol Biol (2024) 31 pp. 1243-1250 [ Pubmed: 38627580 DOI: doi:10.1038/s41594-024-01283-w ]
- Endonuclease gaja (67 kDa, Protein from Bacillus cereus)
- Gabija protein gajb (57 kDa, Protein from Bacillus cereus)
- Tetramer of gabija protein a (Complex from Bacillus cereus)
Structure of Gabija AB complex
Resolution: 3.23 Å
EM Method: Single-particle
Fitted PDBs: 8tk0
Q-score: 0.496
Yang XY, Shen Z, Xie J, Greenwald J, Marathe I, Lin Q, Xie WJ, Wysocki VH, Fu TM
Nat Struct Mol Biol (2024) 31 pp. 1243-1250 [ Pubmed: 38627580 DOI: doi:10.1038/s41594-024-01283-w ]
- Endonuclease gaja (67 kDa, Protein from Bacillus cereus)
- Tetramer of gabija protein a (Complex from Bacillus cereus)
Structure of Gabija AB complex 1
Resolution: 2.98 Å
EM Method: Single-particle
Fitted PDBs: 8tk1
Q-score: 0.453
Yang XY, Shen Z, Xie J, Greenwald J, Marathe I, Lin Q, Xie WJ, Wysocki VH, Fu TM
Nat Struct Mol Biol (2024) 31 pp. 1243-1250 [ Pubmed: 38627580 DOI: doi:10.1038/s41594-024-01283-w ]
- Tetramer of gabija protein ab (4:4) (Complex from Bacillus cereus)
- Endonuclease gaja (67 kDa, Protein from Bacillus cereus)
- Gabija protein gajb (57 kDa, Protein from Bacillus cereus)
Structure of RecT protein from Listeria innoccua phage A118 in complex with 83-mer annealed duplex
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7ub2
Q-score: 0.508
Caldwell BJ, Norris AS, Karbowski CF, Wiegand AM, Wysocki VH, Bell CE
Nat Commun (2022) 13 pp. 7855-7855 [ DOI: doi:10.1038/s41467-022-35572-z Pubmed: 36543802 ]
- Dna (49-mer) (15 kDa, DNA from Escherichia virus M13)
- Dna (49-mer) (14 kDa, DNA from Escherichia virus M13)
- Rect protein from listeria innocua phage a118, complexed with two complementary strands of ssdna that were added to the protein sequentially (602 kDa, Complex from Listeria innocua Clip11262)
- Rect (30 kDa, Protein from Listeria innocua Clip11262)
Structure of RecT protein from Listeria innoccua phage A118 in complex with 83-mer ssDNA
Resolution: 4.5 Å
EM Method: Single-particle
Fitted PDBs: 7ubb
Q-score: 0.166
Caldwell BJ, Norris AS, Karbowski CF, Wiegand AM, Wysocki VH, Bell CE
Nat Commun (2022) 13 pp. 7855-7855 [ DOI: doi:10.1038/s41467-022-35572-z Pubmed: 36543802 ]
Cryo-EM structure of precleavage Cre tetrameric complex
Resolution: 3.23 Å
EM Method: Single-particle
Fitted PDBs: 7rhx
Q-score: 0.526
Stachowski K, Norris AS, Potter D, Wysocki VH, Foster MP
Nucleic Acids Res (2022) 50 pp. 1753-1769 [ Pubmed: 35104890 DOI: doi:10.1093/nar/gkac032 ]
- Dna (42-mer) (13 kDa, DNA from Escherichia phage P1)
- Precleavage synaptic complex of cre recombinase mutant k201a and loxp dna (54 kDa, Complex from Escherichia phage P1)
- Dna (42-mer) (12 kDa, DNA from Escherichia phage P1)
- Recombinase cre (38 kDa, Protein from Escherichia phage P1)
Cre recombinase mutant (D33A/A36V/R192A) in complex with loxA DNA hairpin
Resolution: 3.91 Å
EM Method: Single-particle
Fitted PDBs: 7rhy
Q-score: 0.401
Stachowski K, Norris AS, Potter D, Wysocki VH, Foster MP
Nucleic Acids Res (2022) 50 pp. 1753-1769 [ Pubmed: 35104890 DOI: doi:10.1093/nar/gkac032 ]
- Dna (49-mer) (15 kDa, DNA from Escherichia phage P1)
- Recombinase cre (38 kDa, Protein from Escherichia phage P1)
- Complex of cre recombinase mutant d33a/a36v/r192a and loxa dna hairpin (54 kDa, Complex from Escherichia phage P1)
Resolution: 4.48 Å
EM Method: Single-particle
Fitted PDBs: 7rhz
Q-score: 0.225
Stachowski K, Norris AS, Potter D, Wysocki VH, Foster MP
Nucleic Acids Res (2022) 50 pp. 1753-1769 [ Pubmed: 35104890 DOI: doi:10.1093/nar/gkac032 ]
- Dna (44-mer) (13 kDa, DNA from Escherichia phage P1)
- Heterodimer of cre recombinase mutants d33a/a36v/r192a and r72e/l115d/r119d in complex with loxp dna (100 kDa, Complex from Escherichia phage P1)
- Recombinase cre (38 kDa, Protein from Escherichia phage P1)
- Recombinase cre (38 kDa, Protein from Escherichia phage P1)
- Dna (44-mer) (13 kDa, DNA from Escherichia phage P1)
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 6tu3
Q-score: 0.586
Vimer S, Ben-Nissan G, Morgenstern D, Kumar-Deshmukh F, Polkinghorn C, Quintyn RS, Vasil'ev YV, Beckman JS, Elad N, Wysocki VH, Sharon M
ACS Cent Sci (2020) 6 pp. 573-588 [ DOI: doi:10.1021/acscentsci.0c00080 Pubmed: 32342007 ]
- Proteasome subunit alpha type-3 (28 kDa, Protein from Rattus norvegicus)
- Proteasome subunit beta type-1 (26 kDa, Protein from Rattus norvegicus)
- Rat liver 20s proteasome (Complex from Rattus norvegicus)
- Proteasome subunit beta type-3 (22 kDa, Protein from Rattus norvegicus)
- Proteasome subunit alpha type-7 (28 kDa, Protein from Rattus norvegicus)
- Proteasome subunit beta type-5 (28 kDa, Protein from Rattus norvegicus)
- Proteasome subunit alpha type-5 (26 kDa, Protein from Rattus norvegicus)
- Proteasome subunit beta type-7 (29 kDa, Protein from Rattus norvegicus)
- Proteasome subunit alpha type-1 (29 kDa, Protein from Rattus norvegicus)
- Proteasome subunit beta type-2 (22 kDa, Protein from Rattus norvegicus)
- Proteasome subunit beta type-6 (25 kDa, Protein from Rattus norvegicus)
- Proteasome subunit beta type-4 (29 kDa, Protein from Rattus norvegicus)
- Proteasome subunit alpha type-2 (25 kDa, Protein from Rattus norvegicus)
- Proteasome subunit alpha type-6 (27 kDa, Protein from Rattus norvegicus)
- Proteasome subunit alpha type-4 (29 kDa, Protein from Rattus norvegicus)