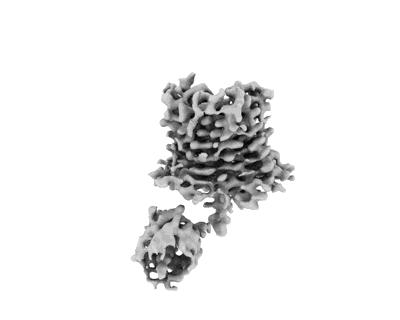
The structure of the core of the pyruvate dehydrogenase complex in the mitochondria of pig hearts.
Cryo-EM map of synthetic cage_T3_5 reconstructed without symmetry (C1)
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
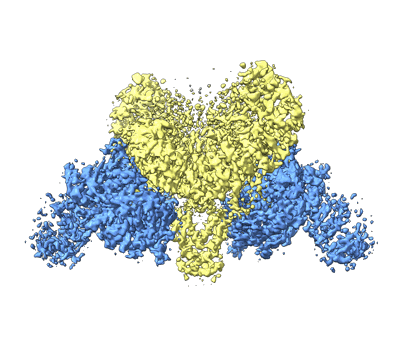
Cryo-EM structure of SARS-CoV-2 Delta variant prefusion S in closed conformation
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 7y6d
Q-score: 0.3
Structure of PfNT1(Y190A)-GFP in complex with GSK4
Resolution: 4.04 Å
EM Method: Single-particle
Fitted PDBs: 7ydq
Q-score: 0.31
CryoEM structure of DDB1-VprBP complex in "ARM-up" conformation
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 7v7b
Q-score: 0.262
To Be Published
- Cryoem structure of ddb1-vprbp complex in "arm-up" conformation (Complex from Homo sapiens)
- Ddb1- and cul4-associated factor 1 (169 kDa, Protein from Homo sapiens)
- Dna damage-binding protein 1 (127 kDa, Protein from Homo sapiens)
Xu J, Ericson CF, Lien YW, Rutaganira FUN, Eisenstein F, Feldmuller M, King N, Pilhofer M
Nat Microbiol (2022) 7 pp. 397-410 [ DOI: doi:10.1038/s41564-022-01059-2 Pubmed: 35165385 ]
Resolution: 4.6 Å
EM Method: Single-particle
Fitted PDBs: 6w5m
Q-score: 0.177
Lee YT, Ayoub A, Park SH, Sha L, Xu J, Mao F, Zheng W, Zhang Y, Cho US, Dou Y
Nat Commun (2021) 12 pp. 2953-2953 [ DOI: doi:10.1038/s41467-021-23268-9 Pubmed: 34012049 ]
- Wd repeat-containing protein 5 (34 kDa, Protein from Homo sapiens)
- Mll1 in complex with rbbp5, wdr5, set1, and ash2l bound to the nucleosome (380 kDa, Complex)
- Dna (Complex from synthetic construct)
- Dna (147-mer) (45 kDa, DNA from synthetic construct)
- Histone h3.2, histone h4, histone h2a type 1, histone h2b 1.1 (Complex from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Retinoblastoma-binding protein 5, wd repeat-containing protein 5, histone-lysine n-methyltransferase 2a, set1/ash2 histone methyltransferase complex subunit ash2 (Complex from Homo sapiens)
- Set1/ash2 histone methyltransferase complex subunit ash2 (60 kDa, Protein from Homo sapiens)
- Dna (147-mer) (45 kDa, DNA from synthetic construct)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Histone h2a type 1 (13 kDa, Protein from Xenopus laevis)
- Histone-lysine n-methyltransferase 2a (24 kDa, Protein from Homo sapiens)
- Retinoblastoma-binding protein 5 (59 kDa, Protein from Homo sapiens)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
Resolution: 4.22 Å
EM Method: Single-particle
Fitted PDBs: 7c17
Q-score: 0.278
Fang C, Philips SJ, Wu X, Chen K, Shi J, Shen L, Xu J, Feng Y, O'Halloran TV, Zhang Y
Nat Chem Biol (2021) 17 pp. 57-64 [ DOI: doi:10.1038/s41589-020-00653-x Pubmed: 32989300 ]
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli (strain K12))
- Dna (72-mer) (22 kDa, DNA from synthetic construct)
- Dna (72-mer) (22 kDa, DNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Silver ion (107 Da, Ligand)
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli (strain K12))
- Rna polymerase sigma factor rpod (72 kDa, Protein from Escherichia coli (strain K12))
- Dna-directed rna polymerase subunit beta' (156 kDa, Protein from Escherichia coli (strain K12))
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli (strain K12))
- Zinc ion (65 Da, Ligand)
- Hth-type transcriptional regulator cuer (15 kDa, Protein from Escherichia coli (strain K12))
- Escherichia coli cuer transcription activation complex with fully duplex promoter dna (507 kDa, Complex from Escherichia coli K-12)
Extended cryo-EM map of native human uromodulin filament core at 4.7 A resolution
Stanisich JJ, Zyla DS, Afanasyev P, Xu J, Kipp A, Olinger E, Devuyst O, Pilhofer M, Boehringer D, Glockshuber R
eLife (2020) 9 [ DOI: doi:10.7554/eLife.60265 Pubmed: 32815518 ]
- Native human uromodulin filament core (1 MDa, Complex from Homo sapiens)
- Native human uromodulin (Protein from Homo sapiens)
cryo-EM structure of alpha2BAR-Gi1 complex
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 6k42
Q-score: 0.299
Yuan D, Liu Z, Kaindl J, Maeda S, Zhao J, Sun X, Xu J, Gmeiner P, Wang HW, Kobilka BK
Nat Chem Biol (2020) 16 pp. 507-512 [ DOI: doi:10.1038/s41589-020-0492-2 Pubmed: 32152538 ]
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (38 kDa, Protein from Mus musculus)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Bos taurus)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Scfv (32 kDa, Protein from Mus musculus)
- 4-[(1~{s})-1-(2,3-dimethylphenyl)ethyl]-1~{h}-imidazole (200 Da, Ligand)
- Alpha2bar-gi1 complex (150 kDa, Complex from Spodoptera)
- Alpha-2a adrenergic receptor,endolysin,alpha-2b adrenergic receptor,alpha-2b adrenergic receptor (58 kDa, Protein from Homo sapiens)