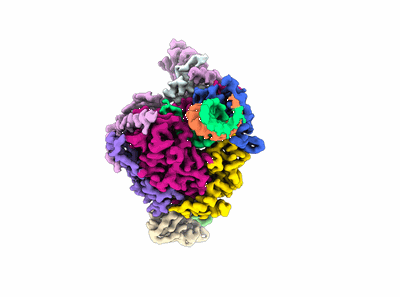
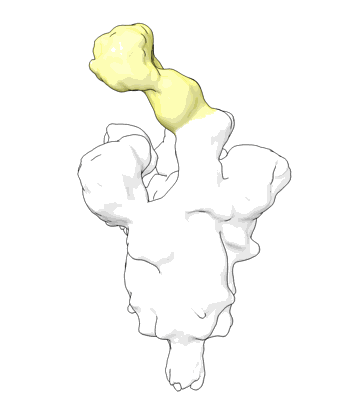
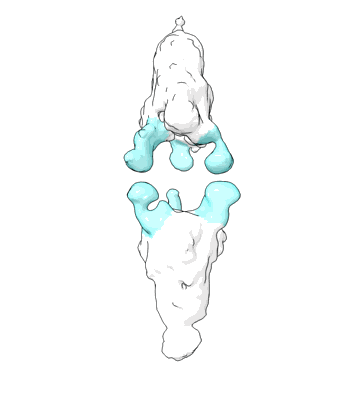
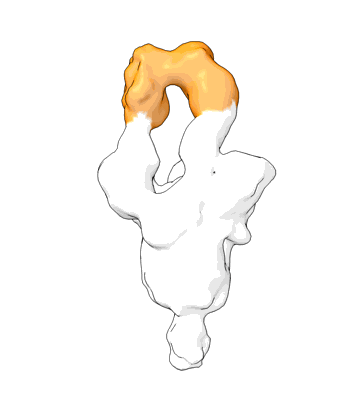
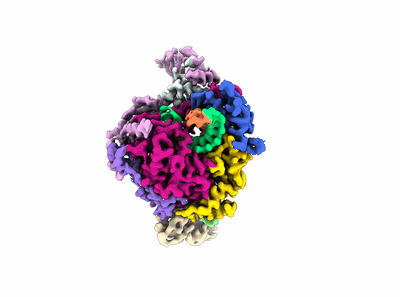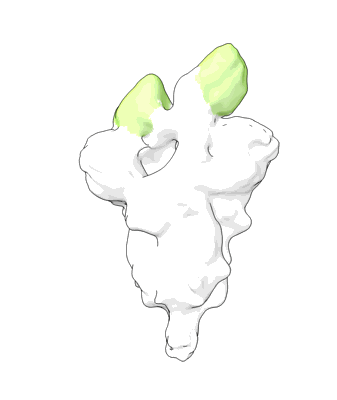Structure of COVID-19 RNA-dependent RNA polymerase bound to penciclovir.
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 7doi
Q-score: 0.599
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Water (18 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Rna (5'-r(p*ap*gp*ap*up*up*ap*ap*gp*up*up*ap*u)-3') (3 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- [(2r)-4-(2-azanyl-6-oxidanylidene-3h-purin-9-yl)-2-(hydroxymethyl)butyl] dihydrogen phosphate (333 Da, Ligand)
- Covid-19 rdrp complex bound to penciclovir (Complex from Severe acute respiratory syndrome coronavirus 2)
- Pyrophosphate 2- (175 Da, Ligand)
- Rna (5'-r(p*cp*cp*up*ap*up*ap*ap*cp*up*up*ap*ap*up*cp*u)-3') (4 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
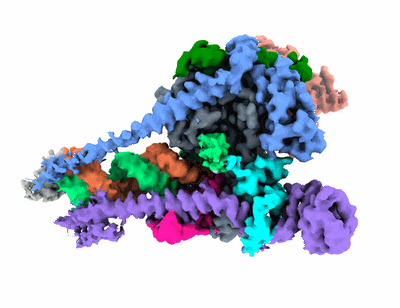
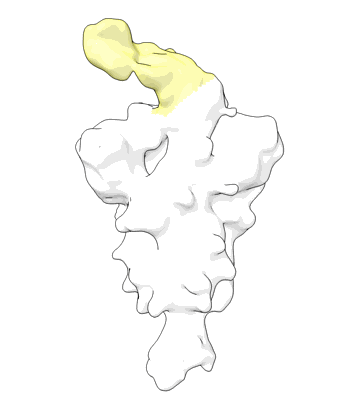
Structure of COVID-19 RNA-dependent RNA polymerase (extended conformation) bound to penciclovir
Resolution: 2.73 Å
EM Method: Single-particle
Fitted PDBs: 7dok
Q-score: 0.554
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Water (18 Da, Ligand)
- Rna-directed rna polymerase (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Rna (5'-r(p*gp*cp*up*ap*up*gp*up*gp*ap*gp*ap*up*up*ap*ap*gp*up*up*ap*u)-3') (6 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- [(2r)-4-(2-azanyl-6-oxidanylidene-3h-purin-9-yl)-2-(hydroxymethyl)butyl] dihydrogen phosphate (333 Da, Ligand)
- Rna (5'-r(p*cp*cp*cp*up*ap*up*ap*ap*cp*up*up*ap*ap*up*cp*up*cp*ap*cp*ap*up*ap*gp*c)-3') (7 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Covid-19 rdrp complex (exntended conformation) bound to penciclovir (Complex from Severe acute respiratory syndrome coronavirus 2)
- Pyrophosphate 2- (175 Da, Ligand)
- Zinc ion (65 Da, Ligand)
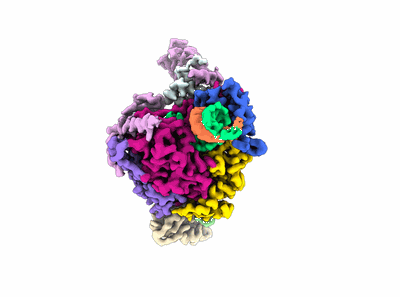
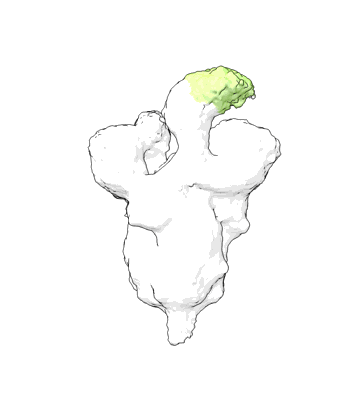
Structure of COVID-19 RNA-dependent RNA polymerase bound to favipiravir
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 7dfg
Q-score: 0.598
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Water (18 Da, Ligand)
- Rna (5'-r(p*cp*cp*cp*up*ap*up*ap*ap*cp*up*up*ap*ap*up*cp*u)-3') (4 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Covid-19 rdrp complex bound to favipiravir (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rna-directed rna polymerase (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Rna (5'-r(p*ap*gp*ap*up*up*ap*ap*gp*up*up*ap*u)-3') (3 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 6-fluoro-3-oxo-4-(5-o-phosphono-beta-d-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide (369 Da, Ligand)
- Pyrophosphate 2- (175 Da, Ligand)
- Zinc ion (65 Da, Ligand)
Structure of COVID-19 RNA-dependent RNA polymerase bound to ribavirin
Resolution: 2.97 Å
EM Method: Single-particle
Fitted PDBs: 7dfh
Q-score: 0.535
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Ribavirin monophosphate (324 Da, Ligand)
- Water (18 Da, Ligand)
- Rna-directed rna polymeras (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (5'-r(p*gp*cp*up*ap*up*gp*up*g*(lig))-3') (2 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Rna (5'-r(p*cp*cp*cp*cp*cp*ap*cp*ap*up*ap*gp*c)-3') (3 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Covid-19 rdrp complex bound to favipiravir (Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Pyrophosphate 2- (175 Da, Ligand)
- Zinc ion (65 Da, Ligand)
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-80 IgG
Hastie KM, Li H, Bedinger D, Schendel SL, Dennison SM, Li K, Rayaprolu V, Yu X, Mann C, Zandonatti M, Diaz Avalos R, Zyla D, Buck T, Hui S, Shaffer K, Hariharan C, Yin J, Olmedillas E, Enriquez A, Parekh D, Abraha M, Feeney E, Horn GQ, Aldon Y, Ali H, Aracic S, Cobb RR, Federman RS, Fernandez JM, Glanville J, Green R, Grigoryan G, Lujan Hernandez AG, Ho DD, Huang KA, Ingraham J, Jiang W, Kellam P, Kim C, Kim M, Kim HM, Kong C, Krebs SJ, Lan F, Lang G, Lee S, Leung CL, Liu J, Lu Y, MacCamy A, McGuire AT, Palser AL, Rabbitts TH, Rikhtegaran Tehrani Z, Sajadi MM, Sanders RW, Sato AK, Schweizer L, Seo J, Shen B, Snitselaar JL, Stamatatos L, Tan Y, Tomic MT, van Gils MJ, Youssef S, Yu J, Yuan TZ, Zhang Q, Peters B, Tomaras GD, Germann T, Saphire EO
Science (2021) 374 pp. 472-478 [ Pubmed: 34554826 DOI: doi:10.1126/science.abh2315 ]
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-081 Fab
Hastie KM, Li H, Bedinger D, Schendel SL, Dennison SM, Li K, Rayaprolu V, Yu X, Mann C, Zandonatti M, Diaz Avalos R, Zyla D, Buck T, Hui S, Shaffer K, Hariharan C, Yin J, Olmedillas E, Enriquez A, Parekh D, Abraha M, Feeney E, Horn GQ, Aldon Y, Ali H, Aracic S, Cobb RR, Federman RS, Fernandez JM, Glanville J, Green R, Grigoryan G, Lujan Hernandez AG, Ho DD, Huang KA, Ingraham J, Jiang W, Kellam P, Kim C, Kim M, Kim HM, Kong C, Krebs SJ, Lan F, Lang G, Lee S, Leung CL, Liu J, Lu Y, MacCamy A, McGuire AT, Palser AL, Rabbitts TH, Rikhtegaran Tehrani Z, Sajadi MM, Sanders RW, Sato AK, Schweizer L, Seo J, Shen B, Snitselaar JL, Stamatatos L, Tan Y, Tomic MT, van Gils MJ, Youssef S, Yu J, Yuan TZ, Zhang Q, Peters B, Tomaras GD, Germann T, Saphire EO
Science (2021) 374 pp. 472-478 [ Pubmed: 34554826 DOI: doi:10.1126/science.abh2315 ]
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-091 IgG
Hastie KM, Li H, Bedinger D, Schendel SL, Dennison SM, Li K, Rayaprolu V, Yu X, Mann C, Zandonatti M, Diaz Avalos R, Zyla D, Buck T, Hui S, Shaffer K, Hariharan C, Yin J, Olmedillas E, Enriquez A, Parekh D, Abraha M, Feeney E, Horn GQ, Aldon Y, Ali H, Aracic S, Cobb RR, Federman RS, Fernandez JM, Glanville J, Green R, Grigoryan G, Lujan Hernandez AG, Ho DD, Huang KA, Ingraham J, Jiang W, Kellam P, Kim C, Kim M, Kim HM, Kong C, Krebs SJ, Lan F, Lang G, Lee S, Leung CL, Liu J, Lu Y, MacCamy A, McGuire AT, Palser AL, Rabbitts TH, Rikhtegaran Tehrani Z, Sajadi MM, Sanders RW, Sato AK, Schweizer L, Seo J, Shen B, Snitselaar JL, Stamatatos L, Tan Y, Tomic MT, van Gils MJ, Youssef S, Yu J, Yuan TZ, Zhang Q, Peters B, Tomaras GD, Germann T, Saphire EO
Science (2021) 374 pp. 472-478 [ Pubmed: 34554826 DOI: doi:10.1126/science.abh2315 ]
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-094 VHH-Fc
Hastie KM, Li H, Bedinger D, Schendel SL, Dennison SM, Li K, Rayaprolu V, Yu X, Mann C, Zandonatti M, Diaz Avalos R, Zyla D, Buck T, Hui S, Shaffer K, Hariharan C, Yin J, Olmedillas E, Enriquez A, Parekh D, Abraha M, Feeney E, Horn GQ, Aldon Y, Ali H, Aracic S, Cobb RR, Federman RS, Fernandez JM, Glanville J, Green R, Grigoryan G, Lujan Hernandez AG, Ho DD, Huang KA, Ingraham J, Jiang W, Kellam P, Kim C, Kim M, Kim HM, Kong C, Krebs SJ, Lan F, Lang G, Lee S, Leung CL, Liu J, Lu Y, MacCamy A, McGuire AT, Palser AL, Rabbitts TH, Rikhtegaran Tehrani Z, Sajadi MM, Sanders RW, Sato AK, Schweizer L, Seo J, Shen B, Snitselaar JL, Stamatatos L, Tan Y, Tomic MT, van Gils MJ, Youssef S, Yu J, Yuan TZ, Zhang Q, Peters B, Tomaras GD, Germann T, Saphire EO
Science (2021) 374 pp. 472-478 [ Pubmed: 34554826 DOI: doi:10.1126/science.abh2315 ]
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-096 IgG
Hastie KM, Li H, Bedinger D, Schendel SL, Dennison SM, Li K, Rayaprolu V, Yu X, Mann C, Zandonatti M, Diaz Avalos R, Zyla D, Buck T, Hui S, Shaffer K, Hariharan C, Yin J, Olmedillas E, Enriquez A, Parekh D, Abraha M, Feeney E, Horn GQ, Aldon Y, Ali H, Aracic S, Cobb RR, Federman RS, Fernandez JM, Glanville J, Green R, Grigoryan G, Lujan Hernandez AG, Ho DD, Huang KA, Ingraham J, Jiang W, Kellam P, Kim C, Kim M, Kim HM, Kong C, Krebs SJ, Lan F, Lang G, Lee S, Leung CL, Liu J, Lu Y, MacCamy A, McGuire AT, Palser AL, Rabbitts TH, Rikhtegaran Tehrani Z, Sajadi MM, Sanders RW, Sato AK, Schweizer L, Seo J, Shen B, Snitselaar JL, Stamatatos L, Tan Y, Tomic MT, van Gils MJ, Youssef S, Yu J, Yuan TZ, Zhang Q, Peters B, Tomaras GD, Germann T, Saphire EO
Science (2021) 374 pp. 472-478 [ Pubmed: 34554826 DOI: doi:10.1126/science.abh2315 ]
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-147 IgG
Hastie KM, Li H, Bedinger D, Schendel SL, Dennison SM, Li K, Rayaprolu V, Yu X, Mann C, Zandonatti M, Diaz Avalos R, Zyla D, Buck T, Hui S, Shaffer K, Hariharan C, Yin J, Olmedillas E, Enriquez A, Parekh D, Abraha M, Feeney E, Horn GQ, Aldon Y, Ali H, Aracic S, Cobb RR, Federman RS, Fernandez JM, Glanville J, Green R, Grigoryan G, Lujan Hernandez AG, Ho DD, Huang KA, Ingraham J, Jiang W, Kellam P, Kim C, Kim M, Kim HM, Kong C, Krebs SJ, Lan F, Lang G, Lee S, Leung CL, Liu J, Lu Y, MacCamy A, McGuire AT, Palser AL, Rabbitts TH, Rikhtegaran Tehrani Z, Sajadi MM, Sanders RW, Sato AK, Schweizer L, Seo J, Shen B, Snitselaar JL, Stamatatos L, Tan Y, Tomic MT, van Gils MJ, Youssef S, Yu J, Yuan TZ, Zhang Q, Peters B, Tomaras GD, Germann T, Saphire EO
Science (2021) 374 pp. 472-478 [ Pubmed: 34554826 DOI: doi:10.1126/science.abh2315 ]