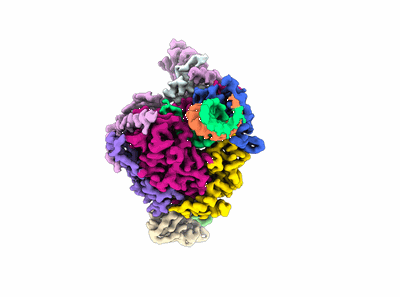
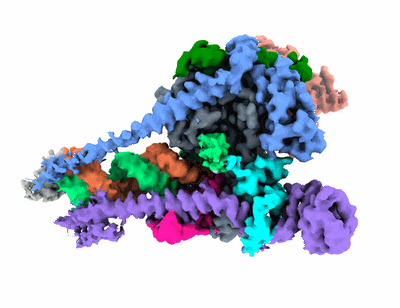
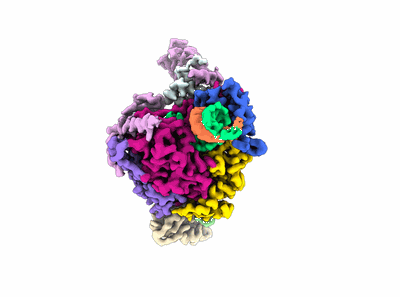
Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab
Resolution: 3.08 Å
EM Method: Single-particle
Fitted PDBs: 7xdl
Q-score: 0.5
Wang Y, Yan A, Song D, Dong C, Rao M, Gao Y, Qi R, Ma X, Wang Q, Xu H, Liu H, Han J, Duan M, Liu S, Yu X, Zong M, Feng J, Jiao J, Zhang H, Li M, Yu B, Wang Y, Meng F, Ni X, Li Y, Shen Z, Sun B, Shao X, Zhao H, Zhao Y, Li R, Zhang Y, Du G, Lu J, You C, Jiang H, Zhang L, Wang L, Dou C, Liu Z, Zhao J
Cell Discov (2023) 9 pp. 3-3 [ Pubmed: 36609558 DOI: doi:10.1038/s41421-022-00509-9 ]
- Cryo-em structure of sars-cov-2 delta spike protein in complex with ba7208 and ba7125 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ba7125 fab (22 kDa, Protein from Homo sapiens)
- Ba7208 and ba7125 fab (Complex)
- Ba7208 fab (11 kDa, Protein from Homo sapiens)
- Spike glycoprotein (123 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ba7208 fab (13 kDa, Protein from Homo sapiens)
- Sars-cov-2 delta spike protein (Complex from Homo sapiens)
- Ba7125 fab (22 kDa, Protein from Homo sapiens)
Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with BA7054 and BA7125 fab (local refinement)
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7xcz
Q-score: 0.528
Wang Y, Yan A, Song D, Dong C, Rao M, Gao Y, Qi R, Ma X, Wang Q, Xu H, Liu H, Han J, Duan M, Liu S, Yu X, Zong M, Feng J, Jiao J, Zhang H, Li M, Yu B, Wang Y, Meng F, Ni X, Li Y, Shen Z, Sun B, Shao X, Zhao H, Zhao Y, Li R, Zhang Y, Du G, Lu J, You C, Jiang H, Zhang L, Wang L, Dou C, Liu Z, Zhao J
Cell Discov (2023) 9 pp. 3-3 [ Pubmed: 36609558 DOI: doi:10.1038/s41421-022-00509-9 ]
- Sars-cov-2 delta rbd (Complex from Homo sapiens)
- Cryo-em structure of sars-cov-2 delta rbd in complex with ba7054 and ba7125 fab (local refinement) (Complex from Severe acute respiratory syndrome coronavirus 2)
- Ba7125 fab (23 kDa, Protein from Homo sapiens)
- Ba7054 fab (23 kDa, Protein from Homo sapiens)
- Ba7054 fab (23 kDa, Protein from Homo sapiens)
- Ba7054 and ba7125 fab (Complex)
- Ba7125 fab (23 kDa, Protein from Homo sapiens)
- Spike glycoprotein (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with BA7208 and BA7125 fab (local refinement)
Resolution: 2.98 Å
EM Method: Single-particle
Fitted PDBs: 7xda
Q-score: 0.526
Wang Y, Yan A, Song D, Dong C, Rao M, Gao Y, Qi R, Ma X, Wang Q, Xu H, Liu H, Han J, Duan M, Liu S, Yu X, Zong M, Feng J, Jiao J, Zhang H, Li M, Yu B, Wang Y, Meng F, Ni X, Li Y, Shen Z, Sun B, Shao X, Zhao H, Zhao Y, Li R, Zhang Y, Du G, Lu J, You C, Jiang H, Zhang L, Wang L, Dou C, Liu Z, Zhao J
Cell Discov (2023) 9 pp. 3-3 [ Pubmed: 36609558 DOI: doi:10.1038/s41421-022-00509-9 ]
- Ba7208 fab (24 kDa, Protein from Homo sapiens)
- Cryo-em structure of sars-cov-2 delta rbd in complex with ba7208 and ba7125 fab (local refinement) (Complex from Homo sapiens)
- Spike glycoprotein (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ba7208 fab (23 kDa, Protein from Homo sapiens)
- Ba7125 fab (23 kDa, Protein from Homo sapiens)
- Ba7125 fab (23 kDa, Protein from Homo sapiens)
Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab
Resolution: 2.62 Å
EM Method: Single-particle
Fitted PDBs: 7xdb
Q-score: 0.502
Wang Y, Yan A, Song D, Dong C, Rao M, Gao Y, Qi R, Ma X, Wang Q, Xu H, Liu H, Han J, Duan M, Liu S, Yu X, Zong M, Feng J, Jiao J, Zhang H, Li M, Yu B, Wang Y, Meng F, Ni X, Li Y, Shen Z, Sun B, Shao X, Zhao H, Zhao Y, Li R, Zhang Y, Du G, Lu J, You C, Jiang H, Zhang L, Wang L, Dou C, Liu Z, Zhao J
Cell Discov (2023) 9 pp. 3-3 [ Pubmed: 36609558 DOI: doi:10.1038/s41421-022-00509-9 ]
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ba7208 fab (11 kDa, Protein from Homo sapiens)
- Sars-cov-2 omicron spike protein (Complex)
- Ba7208 fab (13 kDa, Protein from Homo sapiens)
- Cryo-em structure of sars-cov-2 omicron spike protein in complex with ba7208 fab (Complex from Homo sapiens)
- Ba7208 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7xdk
Q-score: 0.496
Wang Y, Yan A, Song D, Dong C, Rao M, Gao Y, Qi R, Ma X, Wang Q, Xu H, Liu H, Han J, Duan M, Liu S, Yu X, Zong M, Feng J, Jiao J, Zhang H, Li M, Yu B, Wang Y, Meng F, Ni X, Li Y, Shen Z, Sun B, Shao X, Zhao H, Zhao Y, Li R, Zhang Y, Du G, Lu J, You C, Jiang H, Zhang L, Wang L, Dou C, Liu Z, Zhao J
Cell Discov (2023) 9 pp. 3-3 [ Pubmed: 36609558 DOI: doi:10.1038/s41421-022-00509-9 ]
- Spike glycoprotein (123 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ba7054 fab (11 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cryo-em structure of sars-cov-2 delta spike protein in complex with ba7054 and ba7125 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Ba7125 fab (22 kDa, Protein from Homo sapiens)
- Ba7125 fab (23 kDa, Protein from Homo sapiens)
- Sars-cov-2 delta spike protein (Complex from Homo sapiens)
- Ba7054 and ba7125 fab (Complex)
- Ba7054 fab (13 kDa, Protein from Homo sapiens)
Structure of STING SAVI-related mutant V147L
Resolution: 3.65 Å
EM Method: Single-particle
Fitted PDBs: 8gsz
Q-score: 0.445
human STING With agonist HB3089
Resolution: 3.47 Å
EM Method: Single-particle
Fitted PDBs: 8gt6
Q-score: 0.443
Xie Z, Wang Z, Fan F, Zhou J, Hu Z, Wang Q, Wang X, Zeng Q, Zhang Y, Qiu J, Zhou X, Xu H, Bai H, Zhan Z, Ding J, Zhang H, Duan W, Yu X, Geng M
Cell Discov (2022) 8 pp. 133-133 [ DOI: doi:10.1038/s41421-022-00481-4 Pubmed: 36513640 ]
- Human sting with agonist hb3089 (Complex from Homo sapiens)
- Stimulator of interferon genes protein (42 kDa, Protein from Homo sapiens)
- 1-[(2e)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1h-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1h-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1h-pyrazole-5-carbonyl)amino]-7-methyl-1h-furo[3,2-e]benzimidazole-5-carboxamide (873 Da, Ligand)
Structure of COVID-19 RNA-dependent RNA polymerase bound to penciclovir.
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 7doi
Q-score: 0.599
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Rna (5'-r(p*ap*gp*ap*up*up*ap*ap*gp*up*up*ap*u)-3') (3 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Pyrophosphate 2- (175 Da, Ligand)
- Covid-19 rdrp complex bound to penciclovir (Complex from Severe acute respiratory syndrome coronavirus 2)
- [(2r)-4-(2-azanyl-6-oxidanylidene-3h-purin-9-yl)-2-(hydroxymethyl)butyl] dihydrogen phosphate (333 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna-directed rna polymerase (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)
- Rna (5'-r(p*cp*cp*up*ap*up*ap*ap*cp*up*up*ap*ap*up*cp*u)-3') (4 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
Structure of COVID-19 RNA-dependent RNA polymerase (extended conformation) bound to penciclovir
Resolution: 2.73 Å
EM Method: Single-particle
Fitted PDBs: 7dok
Q-score: 0.554
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Rna (5'-r(p*cp*cp*cp*up*ap*up*ap*ap*cp*up*up*ap*ap*up*cp*up*cp*ap*cp*ap*up*ap*gp*c)-3') (7 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Pyrophosphate 2- (175 Da, Ligand)
- [(2r)-4-(2-azanyl-6-oxidanylidene-3h-purin-9-yl)-2-(hydroxymethyl)butyl] dihydrogen phosphate (333 Da, Ligand)
- Covid-19 rdrp complex (exntended conformation) bound to penciclovir (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rna (5'-r(p*gp*cp*up*ap*up*gp*up*gp*ap*gp*ap*up*up*ap*ap*gp*up*up*ap*u)-3') (6 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Rna-directed rna polymerase (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)
Structure of COVID-19 RNA-dependent RNA polymerase bound to favipiravir
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 7dfg
Q-score: 0.598
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Zinc ion (65 Da, Ligand)
- Pyrophosphate 2- (175 Da, Ligand)
- Covid-19 rdrp complex bound to favipiravir (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rna (5'-r(p*cp*cp*cp*up*ap*up*ap*ap*cp*up*up*ap*ap*up*cp*u)-3') (4 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Rna-directed rna polymerase (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (5'-r(p*ap*gp*ap*up*up*ap*ap*gp*up*up*ap*u)-3') (3 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 6-fluoro-3-oxo-4-(5-o-phosphono-beta-d-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide (369 Da, Ligand)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)