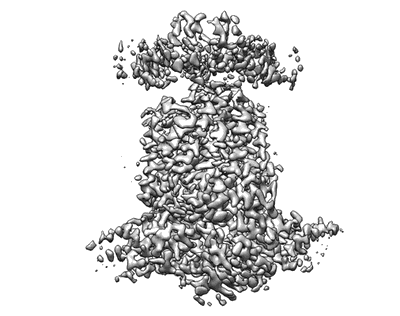
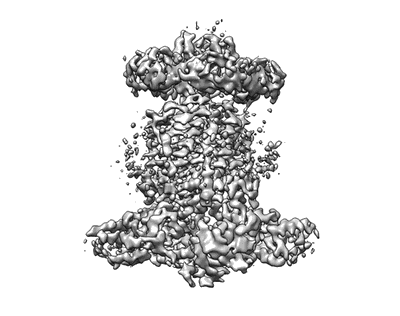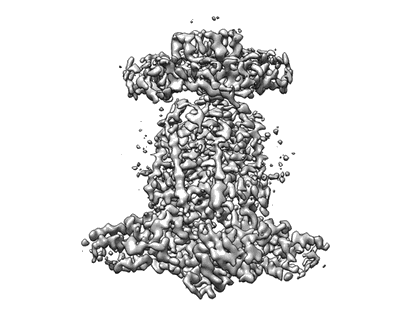Cryo-EM structure of P.aeruginosa MlaFEBD with ADP-V
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 7ch8
Q-score: 0.401
Zhou C, Shi H, Zhang M, Zhou L, Xiao L, Feng S, Im W, Zhou M, Zhang X, Huang Y
J Mol Biol (2021) 433 pp. 166986-166986 [ Pubmed: 33845086 DOI: doi:10.1016/j.jmb.2021.166986 ]
- Probable atp-binding component of abc transporter (29 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- Adp metavanadate (527 Da, Ligand)
- Mlad domain-containing protein (16 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- E.coli mlafeb bond with amppnp (Complex from Escherichia coli K-12)
- Magnesium ion (24 Da, Ligand)
- Stas domain-containing protein (10 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- Probable permease of abc transporter (28 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- 2-(hexadecanoyloxy)-1-[(phosphonooxy)methyl]ethyl hexadecanoate (648 Da, Ligand)
Cryo-EM structure of P.aeruginosa MlaFEBD
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7ch9
Q-score: 0.427
Zhou C, Shi H, Zhang M, Zhou L, Xiao L, Feng S, Im W, Zhou M, Zhang X, Huang Y
J Mol Biol (2021) 433 pp. 166986-166986 [ Pubmed: 33845086 DOI: doi:10.1016/j.jmb.2021.166986 ]
- Cryo-em structure of p.aeruginosa mlafebd (Complex from Pseudomonas aeruginosa PAO1)
- Probable atp-binding component of abc transporter (29 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- Mlad domain-containing protein (16 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- Stas domain-containing protein (10 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- Probable permease of abc transporter (28 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- 2-(hexadecanoyloxy)-1-[(phosphonooxy)methyl]ethyl hexadecanoate (648 Da, Ligand)
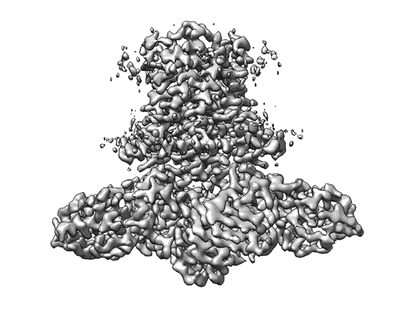
Cryo-EM structure of E.coli MlaFEB with AMPPNP
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7ch6
Q-score: 0.506
Zhou C, Shi H, Zhang M, Zhou L, Xiao L, Feng S, Im W, Zhou M, Zhang X, Huang Y
J Mol Biol (2021) 433 pp. 166986-166986 [ Pubmed: 33845086 DOI: doi:10.1016/j.jmb.2021.166986 ]
- Lipid asymmetry maintenance protein mlab (10 kDa, Protein from Escherichia coli (strain K12))
- Phospholipid abc transporter atp-binding protein mlaf (29 kDa, Protein from Escherichia coli (strain K12))
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
- Lipid asymmetry maintenance abc transporter permease subunit mlae (27 kDa, Protein from Escherichia coli (strain K12))
- E.coli mlafeb bond with amppnp (Complex from Escherichia coli K12)
Cryo-EM structure of E.coli MlaFEB
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 7ch7
Q-score: 0.415
Zhou C, Shi H, Zhang M, Zhou L, Xiao L, Feng S, Im W, Zhou M, Zhang X, Huang Y
J Mol Biol (2021) 433 pp. 166986-166986 [ Pubmed: 33845086 DOI: doi:10.1016/j.jmb.2021.166986 ]
- Lipid asymmetry maintenance protein mlab (10 kDa, Protein from Escherichia coli (strain K12))
- Lipid asymmetry maintenance abc transporter permease subunit mlae (27 kDa, Protein from Escherichia coli (strain K12))
- Phospholipid abc transporter atp-binding protein mlaf (29 kDa, Protein from Escherichia coli (strain K12))
- E.coli mla feb complex (Complex from Escherichia coli K-12)
Cryo-EM structure of P.aeruginosa MlaFEBD with AMPPNP
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 7cha
Q-score: 0.386
Zhou C, Shi H, Zhang M, Zhou L, Xiao L, Feng S, Im W, Zhou M, Zhang X, Huang Y
J Mol Biol (2021) 433 pp. 166986-166986 [ Pubmed: 33845086 DOI: doi:10.1016/j.jmb.2021.166986 ]
- Probable atp-binding component of abc transporter,p.aeruginosa mla f (29 kDa, Protein from Pseudomonas aeruginosa PAO1)
- Mlad domain-containing protein (16 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
- Stas domain-containing protein (10 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- Probable permease of abc transporter (28 kDa, Protein from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1))
- 2-(hexadecanoyloxy)-1-[(phosphonooxy)methyl]ethyl hexadecanoate (648 Da, Ligand)
- Cryo-em structure of p.aeruginosa mlafebd with amppnp (Complex from Pseudomonas aeruginosa PAO1)
Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles
Resolution: 6.0 Å
EM Method: Single-particle
Fitted PDBs: 4au6
Q-score: 0.119
Estrozi LF, Settembre EC, Goret G, McClain B, Zhang X, Chen JZ, Grigorieff N, Harrison SC
J Mol Biol (2013) 425 pp. 124-132 [ Pubmed: 23089332 DOI: doi:10.1016/j.jmb.2012.10.011 ]
- Vp3 (Protein from Bovine rotavirus)
- Vp2 (Protein from Bovine rotavirus)
- Vp1 (Protein from Bovine rotavirus)
- Rotavirus polymerase (vp1) (126 kDa, Protein from Bovine rotavirus)
- Rotavirus dlp+vp1 (Sample)
- Dsrna (RNA from Bovine rotavirus)
Beuron F, Flynn TC, Ma J, Kondo H, Zhang X, Freemont PS
J Mol Biol (2003) 327 pp. 619-629 [ Pubmed: 12634057 ]
- Rat liver endogenous aaa atpase p97 (530 kDa, Sample)
- P97 (97 MDa, Protein from Rattus norvegicus)