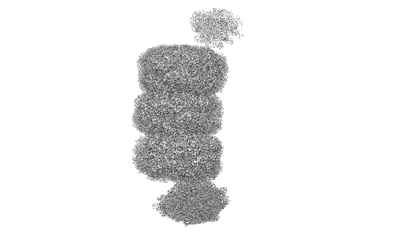
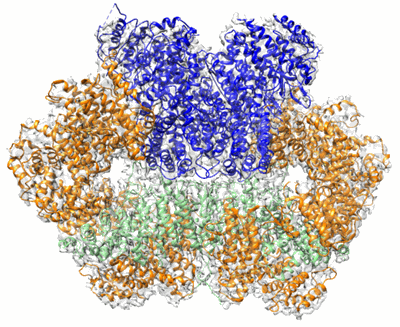
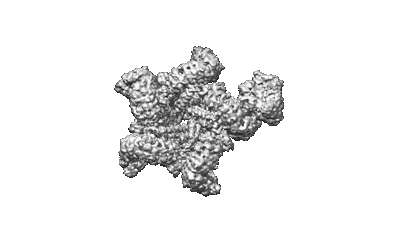
Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 8hfq
Q-score: 0.621
Zheng L, Zhang Z, Wang H, Zheng Z, Wang J, Liu H, Chen H, Dong C, Wang G, Weng Y, Gao N, Zhao J
Nat Commun (2023) 14 pp. 3961-3961 [ Pubmed: 37407580 DOI: doi:10.1038/s41467-023-39689-7 ]
- Photosystem i-associated linker protein cpcl (28 kDa, Protein from Synechocystis sp. (strain PCC 6803 / Kazusa))
- C-phycocyanin alpha subunit (17 kDa, Protein from Synechocystis sp. (strain PCC 6803 / Kazusa))
- Ferredoxin--nadp reductase (46 kDa, Protein from Synechocystis sp. (strain PCC 6803 / Kazusa))
- Phycobilisome 32.1 kda linker polypeptide, phycocyanin-associated, rod 2 (30 kDa, Protein from Synechocystis sp. (strain PCC 6803 / Kazusa))
- Phycobilisome 32.1 kda linker polypeptide, phycocyanin-associated, rod 1 (32 kDa, Protein from Synechocystis sp. (strain PCC 6803 / Kazusa))
- C-phycocyanin beta subunit (18 kDa, Protein from Synechocystis sp. (strain PCC 6803 / Kazusa))
- Cryo-em structure of cpcl-pbs from cyanobacterium synechocystis sp. pcc 6803 (Complex from Synechocystis sp. PCC 6803)
- Phycocyanobilin (588 Da, Ligand)
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 7wzr
Q-score: 0.275
Zhang Q, Wang P, Wu T, Zhang Y, Zheng Z, Zhou S, Qian D, Wang X, Cai G
Cell Discov (2022) 8 pp. 98-98 [ Pubmed: 36175395 DOI: doi:10.1038/s41421-022-00461-8 ]
- Dna damage checkpoint protein lcd1 (86 kDa, Protein from Saccharomyces cerevisiae S288C)
- Serine/threonine-protein kinase mec1 (273 kDa, Protein from Saccharomyces cerevisiae S288C)
- Complex of mec1 and ddc2 (359 kDa, Complex from Saccharomyces cerevisiae)
Cryo-EM structure of MEC1-DDC2-MMS
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 7wzw
Q-score: 0.227
Zhang Q, Wang P, Wu T, Zhang Y, Zheng Z, Zhou S, Qian D, Wang X, Cai G
Cell Discov (2022) 8 pp. 98-98 [ Pubmed: 36175395 DOI: doi:10.1038/s41421-022-00461-8 ]
- Dna damage checkpoint protein lcd1 (86 kDa, Protein from Saccharomyces cerevisiae S288C)
- Mec1-ddc2 (Complex from Saccharomyces cerevisiae)
- Serine/threonine-protein kinase mec1 (273 kDa, Protein from Saccharomyces cerevisiae S288C)
Cryo-EM structure of Sr35 resistosome induced by AvrSr35 R381A
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7xx2
Q-score: 0.418
Zhao YB, Liu MX, Chen TT, Ma X, Li ZK, Zheng Z, Zheng SR, Chen L, Li YZ, Tang LR, Chen Q, Wang P, Ouyang S
Sci Adv (2022) 8 pp. eabq5108-eabq5108 [ Pubmed: 36083908 DOI: doi:10.1126/sciadv.abq5108 ]
- Cnl9 (105 kDa, Protein from Triticum monococcum)
- Sr35 resistosome (Complex)
- Avrsr35 (Complex from Puccinia graminis f.sp.tritici)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Sr35 (Complex from Triticum monococcum)
- Avrsr35 (66 kDa, Protein from Puccinia graminis f. sp. tritici)
Cryo-EM structure of plant NLR Sr35 resistosome
Resolution: 3.33 Å
EM Method: Single-particle
Fitted PDBs: 7xe0
Q-score: 0.351
Zhao YB, Liu MX, Chen TT, Ma X, Li ZK, Zheng Z, Zheng SR, Chen L, Li YZ, Tang LR, Chen Q, Wang P, Ouyang S
Sci Adv (2022) 8 pp. eabq5108-eabq5108 [ Pubmed: 36083908 DOI: doi:10.1126/sciadv.abq5108 ]
- Avrsr35 (Complex from Puccinia graminis f. sp. tritici)
- Sr35 resistosome (Complex)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Sr35 (105 kDa, Protein from Triticum monococcum)
- Sr35 (Complex from Triticum monococcum)
- Avrsr35 (66 kDa, Protein from Puccinia graminis f. sp. tritici)