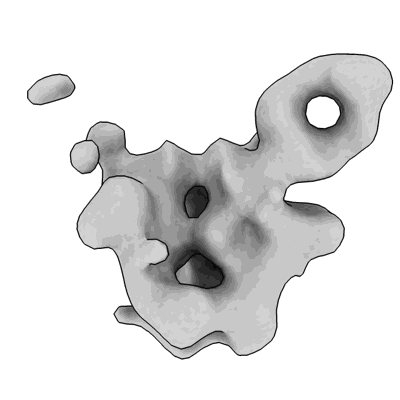
Structure of a native Drosophila melanogaster octameric nucleosome
Resolution: 3.29 Å
EM Method: Single-particle
Fitted PDBs: 9mu4
Q-score: 0.442
Venette-Smith NL, Vishwakarma RK, Dollinger R, Schultz J, Venkatakrishnan V, Babitzke P, Anand G, Gilmour DS, Armache J-P, Murakami K
To Be Published
- Dna (164-mer) (50 kDa, DNA from Drosophila melanogaster)
- Histone h2a (11 kDa, Protein from Drosophila melanogaster)
- Histone h4 (9 kDa, Protein from Drosophila melanogaster)
- Histone h3 (11 kDa, Protein from Drosophila melanogaster)
- Dna (164-mer) (50 kDa, DNA from Drosophila melanogaster)
- Histone h2b (10 kDa, Protein from Drosophila melanogaster)
- Natively purified octameric nucleosome from drosophila melanogaster (240 MDa, Complex from Drosophila melanogaster)
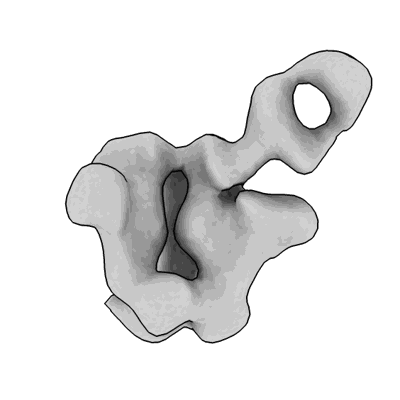
Structure of a native Drosophila melanogaster hexameric nucleosome
Resolution: 6.3 Å
EM Method: Single-particle
Fitted PDBs: 9mu5
Q-score: 0.206
Venette-Smith NL, Vishwakarma RK, Dollinger R, Schultz J, Venkatakrishnan V, Babitzke P, Anand G, Gilmour DS, Armache J-P, Murakami K
To Be Published
- Natively purified hexameric nucleosome from drosophila melanogaster (220 MDa, Complex from Drosophila melanogaster)
- Dna (133-mer) (41 kDa, DNA from Drosophila melanogaster)
- Histone h4 (9 kDa, Protein from Drosophila melanogaster)
- Histone h2b (10 kDa, Protein from Drosophila melanogaster)
- Histone h3 (10 kDa, Protein from Drosophila melanogaster)
- Dna (133-mer) (40 kDa, DNA from Drosophila melanogaster)
- Histone h2a (11 kDa, Protein from Drosophila melanogaster)
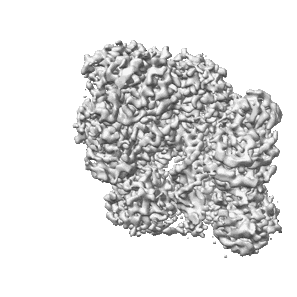
Resolution: 7.8 Å
EM Method: Single-particle
Fitted PDBs: 9mu9
Q-score: 0.202
Venette-Smith NL, Vishwakarma RK, Dollinger R, Schultz J, Venkatakrishnan V, Babitzke P, Anand G, Gilmour DS, Armache J-P, Murakami K
To Be Published
- Non-template dna (50 kDa, DNA from Drosophila melanogaster)
- Rna (3 kDa, RNA from Drosophila melanogaster)
- Template dna (53 kDa, DNA from Drosophila melanogaster)
- Histone h3 (11 kDa, Protein from Drosophila melanogaster)
- Histone h4 (9 kDa, Protein from Drosophila melanogaster)
- Dna-directed rna polymerase ii subunit rpb11 (13 kDa, Protein from Drosophila melanogaster)
- Dna-directed rna polymerase ii subunit rpb5 (24 kDa, Protein from Drosophila melanogaster)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc2 (9 kDa, Protein from Drosophila melanogaster)
- Dna-directed rna polymerase ii subunit rpb3 (30 kDa, Protein from Drosophila melanogaster)
- Zinc ion (65 Da, Ligand)
- Histone h2b (10 kDa, Protein from Drosophila melanogaster)
- Dna-directed rna polymerase ii subunit rpb12 (5 kDa, Protein from Drosophila melanogaster)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (16 kDa, Protein from Drosophila melanogaster)
- Native purified nucleosome elongation complex from drosophila melanogaster (750 kDa, Complex from Drosophila melanogaster)
- Histone h2a (11 kDa, Protein from Drosophila melanogaster)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (7 kDa, Protein from Drosophila melanogaster)
- Dna-directed rna polymerase ii subunit rpb2 (132 kDa, Protein from Drosophila melanogaster)
- Dna-directed rna polymerase ii subunit rpb1 (166 kDa, Protein from Drosophila melanogaster)
- Dna-directed rna polymerase ii subunit rpb9 (13 kDa, Protein from Drosophila melanogaster)
Chao CW, Sprouse KR, Miranda MC, Catanzaro NJ, Hubbard ML, Addetia A, Stewart C, Brown JT, Dosey A, Valdez A, Ravichandran R, Hendricks GG, Ahlrichs M, Dobbins C, Hand A, McGowan J, Simmons B, Treichel C, Willoughby I, Walls AC, McGuire AT, Leaf EM, Baric RS, Schafer A, Veesler D, King NP
Cell Rep (2024) 43 pp. 115036-115036 [ DOI: doi:10.1016/j.celrep.2024.115036 Pubmed: 39644492 ]
sRT-V3 NFL trimer in complex with 12CA5 Fab and RM20A3 Fab
To Be Published
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8pnf
Q-score: 0.601
Gil-Cantero D, Mata CP, Valiente L, Rodriguez-Huete A, Valbuena A, Twarock R, Stockley PG, Mateu MG, Caston JR
Commun Biol (2024) 7 pp. 1501-1501 [ DOI: doi:10.1038/s42003-024-07213-2 Pubmed: 39537894 ]
- Capsid protein vp1 (31 kDa, Protein from rhinovirus B14)
- Capsid protein vp3 (26 kDa, Protein from rhinovirus B14)
- Capsid protein vp2 (27 kDa, Protein from rhinovirus B14)
- Capsid protein vp4 (5 kDa, Protein from rhinovirus B14)
- Rna (5'-r(p*ap*up*up*up*up*up*up*up*up*up*up*up*up*u)-3') (4 kDa, RNA from rhinovirus B14)
- Rhinovirus b14 (Virus from rhinovirus B14)
Xie Z, Lin YC, Steichen JM, Ozorowski G, Kratochvil S, Ray R, Torres JL, Liguori A, Kalyuzhniy O, Wang X, Warner JE, Weldon SR, Dale GA, Kirsch KH, Nair U, Baboo S, Georgeson E, Adachi Y, Kubitz M, Jackson AM, Richey ST, Volk RM, Lee JH, Diedrich JK, Prum T, Falcone S, Himansu S, Carfi A, Yates 3rd JR, Paulson JC, Sok D, Ward AB, Schief WR, Batista FD
Science (2024) 384 pp. eadk0582-eadk0582 [ DOI: doi:10.1126/science.adk0582 Pubmed: 38753770 ]
Xie Z, Lin YC, Steichen JM, Ozorowski G, Kratochvil S, Ray R, Torres JL, Liguori A, Kalyuzhniy O, Wang X, Warner JE, Weldon SR, Dale GA, Kirsch KH, Nair U, Baboo S, Georgeson E, Adachi Y, Kubitz M, Jackson AM, Richey ST, Volk RM, Lee JH, Diedrich JK, Prum T, Falcone S, Himansu S, Carfi A, Yates 3rd JR, Paulson JC, Sok D, Ward AB, Schief WR, Batista FD
Science (2024) 384 pp. eadk0582-eadk0582 [ DOI: doi:10.1126/science.adk0582 Pubmed: 38753770 ]
Structure of the E. coli clamp loader bound to the beta clamp in a Open-DNAp/t conformation
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 8val
Q-score: 0.434
Landeck JT, Pajak J, Norman EK, Sedivy EL, Kelch BA
J Biol Chem (2024) 300 pp. 107166-107166 [ DOI: doi:10.1016/j.jbc.2024.107166 Pubmed: 38490435 ]
- Dna polymerase iii subunit delta (38 kDa, Protein from Escherichia coli)
- Dna (5'-d(p*tp*tp*tp*tp*tp*tp*tp*ap*tp*gp*tp*ap*cp*tp*cp*gp*tp*ap*gp*tp*gp*tp*cp*t)-3') (9 kDa, DNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Dna polymerase iii subunit tau (41 kDa, Protein from Escherichia coli)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Beryllium trifluoride ion (66 Da, Ligand)
- Dna polymerase iii subunit delta' (37 kDa, Protein from Escherichia coli)
- Zinc ion (65 Da, Ligand)
- Structure of the e. coli clamp loader bound to the beta clamp in an open-dnap/t conformation (Complex from Escherichia coli)
- Beta sliding clamp (40 kDa, Protein from Escherichia coli)
- Dna (5'-d(*ap*gp*ap*cp*ap*cp*tp*ap*cp*gp*ap*gp*tp*ap*cp*ap*tp*a)-3') (6 kDa, DNA from synthetic construct)
Structure of the E. coli clamp loader bound to the beta clamp in a Closed-DNA1 conformation
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 8vaq
Q-score: 0.421
Landeck JT, Pajak J, Norman EK, Sedivy EL, Kelch BA
J Biol Chem (2024) 300 pp. 107166-107166 [ DOI: doi:10.1016/j.jbc.2024.107166 Pubmed: 38490435 ]
- Dna polymerase iii subunit delta (38 kDa, Protein from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Dna polymerase iii subunit tau (41 kDa, Protein from Escherichia coli)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Beryllium trifluoride ion (66 Da, Ligand)
- Dna polymerase iii subunit delta' (37 kDa, Protein from Escherichia coli)
- Dna (5'-d(p*gp*cp*ap*gp*ap*cp*ap*cp*tp*ap*cp*gp*ap*gp*tp*ap*cp*ap*tp*a)-3') (6 kDa, DNA from synthetic construct)
- Dna (27-mer) (9 kDa, DNA from synthetic construct)
- Zinc ion (65 Da, Ligand)
- Structure of the e. coli clamp loader bound to the beta clamp in a closed-dna1 conformation (Complex from Escherichia coli)
- Beta sliding clamp (40 kDa, Protein from Escherichia coli)