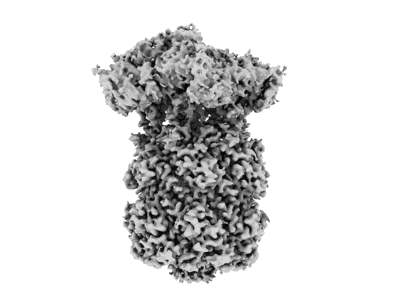
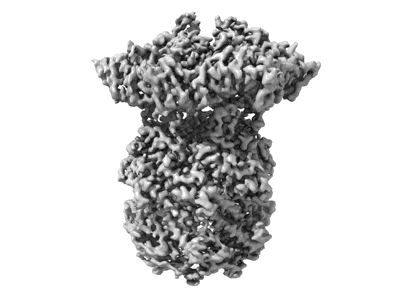
CryoEM structure of apo M. tuberculosis ClpP1P2
Zhou B, Gao Y, Zhao H, Chen X, He J, Zhang T, Xiong X
To Be Published
- Atp-dependent clp protease proteolytic subunit 1 (21 kDa, Protein from Mycobacterium tuberculosis H37Rv)
- Mycobacterium tuberculosis caseinolytic protease p1p2 (Complex from Mycobacterium tuberculosis)
- Atp-dependent clp protease proteolytic subunit 2 (23 kDa, Protein from Mycobacterium tuberculosis H37Rv)
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8v9r
Q-score: 0.475
Ghanbarpour A, Sauer RT, Davis JH
To Be Published
- Atp-dependent clp protease atp-binding subunit clpx (42 kDa, Protein from Escherichia coli)
- Dihydrofolate reductase (23 kDa, Protein from Escherichia coli)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Methotrexate (454 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Atp-dependent clp protease proteolytic subunit (23 kDa, Protein from Escherichia coli)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Clpx.clpp.dhfr (Complex from Escherichia coli)
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 9c87
Q-score: 0.335
Ghanbarpour A, Sauer RT, Davis JH
To Be Published
- Clpx.clpp.linear-degron dhfr-ssra (Complex from Escherichia coli)
- Atp-dependent clp protease proteolytic subunit (23 kDa, Protein from Escherichia coli)
- Dihydrofolate reductase (23 kDa, Protein from Escherichia coli)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Atp-dependent clp protease atp-binding subunit clpx (42 kDa, Protein from Escherichia coli)
- Methotrexate (454 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 9c88
Q-score: 0.5
Ghanbarpour A, Sauer RT, Davis JH
To Be Published
- Atp-dependent clp protease atp-binding subunit clpx (42 kDa, Protein from Escherichia coli)
- Branched-degron dhfr (869 Da, Protein from Escherichia coli)
- Atp-dependent clp protease proteolytic subunit (23 kDa, Protein from Escherichia coli)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Clpx.clpp.dhfr (Complex from Escherichia coli)
Cryo-EM structure of the ClpP degradation system in Streptomyces hawaiiensis
Resolution: 2.34 Å
EM Method: Single-particle
Fitted PDBs: 8xn4
Q-score: 0.56
Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis
Resolution: 1.96 Å
EM Method: Single-particle
Fitted PDBs: 8xon
Q-score: 0.503
Xu X, Wang Y, Huang W, Li D, Deng Z, Long F
Mbio (2024) 15 pp. e0003124-e0003124 [ Pubmed: 38501868 DOI: doi:10.1128/mbio.00031-24 ]
- Ndp-hexose 4-ketoreductase (77 kDa, Protein from Streptomyces hawaiiensis)
- Clpp1,clpp2,clpc1,casein (Complex from Streptomyces hawaiiensis)
- Atp-dependent clp protease proteolytic subunit (24 kDa, Protein from Streptomyces hawaiiensis)
- Atp-dependent clp protease proteolytic subunit (22 kDa, Protein from Streptomyces hawaiiensis)
- Casein (2 kDa, Protein from Bos taurus)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis
Resolution: 1.84 Å
EM Method: Single-particle
Fitted PDBs: 8xoo
Q-score: 0.514
Xu X, Wang Y, Huang W, Li D, Deng Z, Long F
Mbio (2024) 15 pp. e0003124-e0003124 [ Pubmed: 38501868 DOI: doi:10.1128/mbio.00031-24 ]
- Ndp-hexose 4-ketoreductase (77 kDa, Protein from Streptomyces hawaiiensis)
- Clpp1,clpp2,clpc1,casein (Complex from Streptomyces hawaiiensis)
- Atp-dependent clp protease proteolytic subunit (24 kDa, Protein from Streptomyces hawaiiensis)
- Atp-dependent clp protease proteolytic subunit (22 kDa, Protein from Streptomyces hawaiiensis)
- Casein (2 kDa, Protein from Bos taurus)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
Cryo-EM structure of ClpP1P2 in complex with ADEP1 from Streptomyces hawaiiensis
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8xop
Q-score: 0.498
Xu X, Wang Y, Huang W, Li D, Deng Z, Long F
Mbio (2024) 15 pp. e0003124-e0003124 [ Pubmed: 38501868 DOI: doi:10.1128/mbio.00031-24 ]
- Adep1 (736 Da, Protein from synthetic construct)
- Atp-dependent clp protease proteolytic subunit (24 kDa, Protein from Streptomyces hawaiiensis)
- Atp-dependent clp protease proteolytic subunit (22 kDa, Protein from Streptomyces hawaiiensis)
- Clpp1,clpp2 (Complex from Streptomyces hawaiiensis)
Cryo-EM structure of substrate-free DNClpX.ClpP from singly capped particles
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8e7v
Q-score: 0.468
Ghanbarpour A, Cohen S, Davis JH, Sauer RT
Nat Commun (2023) [ ]
- Atp-dependent clp protease atp-binding subunit clpx (42 kDa, Protein from Escherichia coli)
- Clpxp aaa protease (Complex from Escherichia coli)
- Atp-dependent clp protease proteolytic subunit (23 kDa, Protein from Escherichia coli)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
Cryo-EM structure of substrate-free DNClpX.ClpP
Resolution: 3.12 Å
EM Method: Single-particle
Fitted PDBs: 8e8q
Q-score: 0.452
Ghanbarpour A, Cohen S, Davis JH, Sauer RT
Nat Commun (2023) [ ]
- Atp-dependent clp protease atp-binding subunit clpx (42 kDa, Protein from Escherichia coli)
- Clpxp aaa + protease (Complex from Escherichia coli)
- Atp-dependent clp protease proteolytic subunit (23 kDa, Protein from Escherichia coli)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)