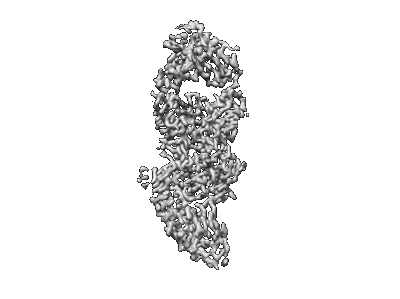
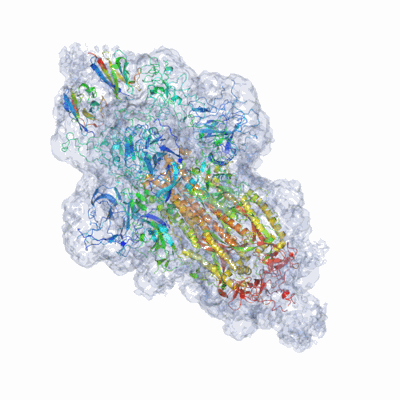
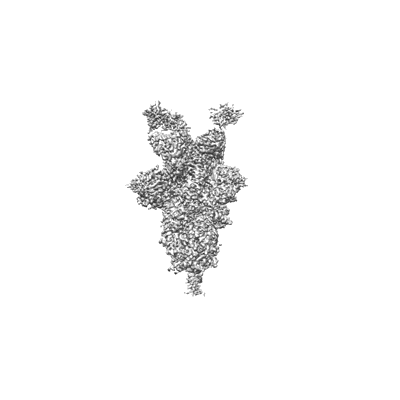
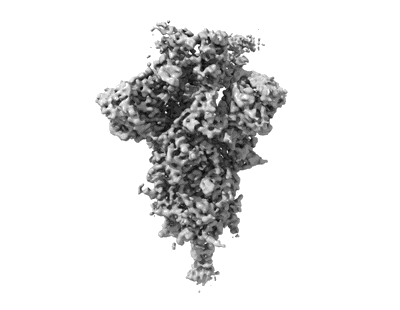
SARS-CoV-2 spike omicron (BA.1) ectodomain trimer in complex with SC27 Fab, local refinement
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8vif
Q-score: 0.545
Voss WN, Mallory MA, Byrne PO, Marchioni JM, Knudson SA, Powers JM, Leist SR, Dadonaite B, Townsend DR, Kain J, Huang Y, Satterwhite E, Castillo IN, Mattocks M, Paresi C, Munt JE, Scobey T, Seeger A, Premkumar L, Bloom JD, Georgiou G, McLellan JS, Baric RS, Lavinder JJ, Ippolito GC
Cell Rep Med (2024) 5 pp. 101668-101668 [ Pubmed: 39094579 DOI: doi:10.1016/j.xcrm.2024.101668 ]
- Sc27 fab (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike omicron (ba.1) ectodomain (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike omicron (ba.1) ectodomain trimer in complex with sc27 fab, global refinement (Complex)
- Sc27 fab heavy chain (14 kDa, Protein from Homo sapiens)
- Sc27 fab light chain (11 kDa, Protein from Homo sapiens)
- Spike glycoprotein (144 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 spike omicron (BA.1) RBD ectodomain dimer-of-trimers in complex with SC27 Fabs
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8vke
Q-score: 0.544
Voss WN, Mallory MA, Byrne PO, Marchioni JM, Knudson SA, Powers JM, Leist SR, Dadonaite B, Townsend DR, Kain J, Huang Y, Satterwhite E, Castillo IN, Mattocks M, Paresi C, Munt JE, Scobey T, Seeger A, Premkumar L, Bloom JD, Georgiou G, McLellan JS, Baric RS, Lavinder JJ, Ippolito GC
Cell Rep Med (2024) 5 pp. 101668-101668 [ Pubmed: 39094579 DOI: doi:10.1016/j.xcrm.2024.101668 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sc27 fab light chain (25 kDa, Protein from Homo sapiens)
- Sc27 fab heavy chain (27 kDa, Protein from Homo sapiens)
- Spike glycoprotein (144 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike omicron (ba.1) rbd ectodomain dimer-of-trimers in complex with sc27 fabs (Complex)
Resolution: 2.83 Å
EM Method: Single-particle
Fitted PDBs: 8thf
Q-score: 0.461
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 8oyt
Q-score: 0.161
Cornish K, Huo J, Jones L, Sharma P, Thrush JW, Abdelkarim S, Kipar A, Ramadurai S, Weckener M, Mikolajek H, Liu S, Buckle I, Bentley E, Kirby A, Han X, Laidlaw SM, Hill M, Eyssen L, Norman C, Le Bas A, Clarke J, James W, Stewart JP, Carroll M, Naismith JH, Owens RJ
Open Biol (2024) 14 pp. 230252-230252 [ DOI: doi:10.1098/rsob.230252 Pubmed: 38835241 ]
- H6 nanobody (Complex from Lama glama)
- Stabilised hexapro omicron ba.1 spike trimer (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- H6 nanobody (14 kDa, Protein from Lama glama)
- Spike glycoprotein,fibritin (140 kDa, Protein from Tequatrovirus T4)
- Complex of stabilised hexapro omicron ba.1 spike trimer with three h6 nanobodies (Complex)
SARS-CoV2 Omicron BA.1 spike in complex with CAB-A17 antibody
Resolution: 2.56 Å
EM Method: Single-particle
Fitted PDBs: 8c2r
Q-score: 0.436
Sheward DJ, Pushparaj P, Das H, Greaney AJ, Kim C, Kim S, Hanke L, Hyllner E, Dyrdak R, Lee J, Dopico XC, Dosenovic P, Peacock TP, McInerney GM, Albert J, Corcoran M, Bloom JD, Murrell B, Karlsson Hedestam GB, Hallberg BM
Cell Rep Med (2024) 5 pp. 101577-101577 [ DOI: doi:10.1016/j.xcrm.2024.101577 Pubmed: 38761799 ]
- Cab-a17 antibody (lc) (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cab-a17 antibody (fab) (Complex from Enterobacteria phage T4)
- Sars-cov2 omicron ba.1 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov2 omicron ba.1 spike in complex with cab-a17 antibody (95 kDa, Complex)
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Cab-a17 antibody (hc) (22 kDa, Protein from Homo sapiens)
Liu B, Liu H, Han P, Wang X, Wang C, Yan X, Lei W, Xu K, Zhou J, Qi J, Fan R, Wu G, Tian WX, Gao GF, Wang Q
Signal Transduct Target Ther (2024) 9 pp. 131-131 [ Pubmed: 38740785 DOI: doi:10.1038/s41392-024-01847-8 ]
- Nanobody n235 (13 kDa, Protein from Vicugna pacos)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- S2l20 heavy chain (52 kDa, Protein from Homo sapiens)
- The n-terminal domain of omicron ba.1 in complex with nanobody n235 and s2l20 fab (Complex from Vicugna pacos)
- S2l20 light chain (26 kDa, Protein from Homo sapiens)
- Spike protein s2' (34 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 8oyu
Q-score: 0.145
Cornish K, Huo J, Jones L, Sharma P, Thrush JW, Abdelkarim S, Kipar A, Ramadurai S, Weckener M, Mikolajek H, Liu S, Buckle I, Bentley E, Kirby A, Han X, Laidlaw SM, Hill M, Eyssen L, Norman C, Le Bas A, Clarke J, James W, Stewart JP, Carroll M, Naismith JH, Owens RJ
Open Biol (2024) 14 pp. 230252-230252 [ DOI: doi:10.1098/rsob.230252 Pubmed: 38835241 ]
- H6 nanobody (Complex from Lama glama)
- Stabilised hexapro omicron ba.1 spike trimer (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Complex of stabilised hexapro omicron ba.1 spike trimer with two h6 nanobodies (Complex)
- Spike glycoprotein,fibritin (139 kDa, Protein from Enterobacteria phage T4)
- H6 nanobody (13 kDa, Protein from Lama glama)
SARS-CoV-2 Omicron BA.1 spike trimer (x2-4P) in complex with 3 D1F6 Fabs (0 RBD up)
Resolution: 3.67 Å
EM Method: Single-particle
Fitted PDBs: 8zby
Q-score: 0.372
Liu B, Niu X, Deng Y, Zhang Z, Wang Y, Gao X, Liang H, Li Z, Wang Q, Cheng Y, Chen Q, Huang S, Pan Y, Su M, Lin X, Niu C, Chen Y, Yang W, Zhang Y, Yan Q, He J, Zhao J, Chen L, Xiong X
Cell Rep (2024) 43 pp. 114265-114265 [ DOI: doi:10.1016/j.celrep.2024.114265 Pubmed: 38805396 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Light chain of d1f6 fab (23 kDa, Protein from Homo sapiens)
- Sars-cov-2 omicron ba.1 spike trimer (x2-4p) in complex with 3 d1f6 fabs (0 rbd up) (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (138 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Heavy chain of d1f6 fab (24 kDa, Protein from Homo sapiens)
SARS-CoV-2 BA.1 spike with UT28-RD
Resolution: 3.22 Å
EM Method: Single-particle
Fitted PDBs: 8k5h
Q-score: 0.497
Ozawa T, Ikeda Y, Chen L, Suzuki R, Hoshino A, Noguchi A, Kita S, Anraku Y, Igarashi E, Saga Y, Inasaki N, Taminishi S, Sasaki J, Kirita Y, Fukuhara H, Maenaka K, Hashiguchi T, Fukuhara T, Hirabayashi K, Tani H, Kishi H, Niimi H
Structure (2024) 32 pp. 263-272.e7 [ DOI: doi:10.1016/j.str.2023.12.013 Pubmed: 38228146 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ut28k-rd fab heavy chain (27 kDa, Protein from Homo sapiens)
- Sars-cov-2 ba.1 spike protein with ut28k-rd complex (50 kDa, Complex)
- Ut28k-rd fab light chain (25 kDa, Protein from Homo sapiens)
- Spike glycoprotein (133 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ut28k-rd fab (Complex from Homo sapiens)
- Sars-cov-2 ba.1 spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
Resolution: 3.66 Å
EM Method: Single-particle
Fitted PDBs: 8k45
Q-score: 0.408
Yao H, Wang H, Zhang Z, Lu Y, Zhang Z, Zhang Y, Xiong X, Wang Y, Wang Z, Yang H, Zhao J, Xu W
MedComm (2020) (2023) 4 pp. e397-e397 [ Pubmed: 37901798 DOI: doi:10.1002/mco2.397 ]
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Nb4 nanobody (13 kDa, Protein from Vicugna pacos)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 omicron ba.1 spike glycoprotein complex with nb4 nanobody (Complex from Severe acute respiratory syndrome coronavirus 2)