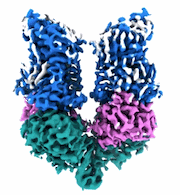
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with GABA and puerarin
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 9eqg
Q-score: 0.627
Lyu Q, Xue W, Liu R, Ma Q, Kasaragod VB, Sun S, Li Q, Chen Y, Yuan M, Yang Y, Zhang B, Nie A, Jia S, Shen C, Gao P, Rong W, Yu C, Bi Y, Zhang C, Nan F, Ning G, Rao Z, Yang X, Wang J, Wang W
Nature (2024) 634 pp. 936-943 [ Pubmed: 39261733 DOI: doi:10.1038/s41586-024-07929-5 ]
- Gamma-aminobutyric acid receptor subunit beta-3 (54 kDa, Protein from Homo sapiens)
- Hexane (86 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 1,2-dilauroyl-sn-glycero-3-phosphate (535 Da, Ligand)
- Water (18 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Gamma-amino-butanoic acid (103 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit gamma-2 (56 kDa, Protein from Homo sapiens)
- [(2r)-1-octadecanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8z)-icosa-5,8,11,14-tetraenoate (1 kDa, Ligand)
- Hexadecane (226 Da, Ligand)
- Decane (142 Da, Ligand)
- (1r)-2-{[(s)-{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9z)-octadec-9-enoate (749 Da, Ligand)
- Puerarin (416 Da, Ligand)
- Cryoem structure of human full-length alpha1beta3gamma2l gaba(a)r in complex with gaba and puerarin (250 kDa, Complex from Homo sapiens)
- Chloride ion (35 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit alpha-1 (52 kDa, Protein from Homo sapiens)
Human EBP complexed with compound 3a
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8w0s
Q-score: 0.558
Dorel R, Sun D, Carruthers N, Castanedo GM, Ung PM, Factor DC, Li T, Baumann H, Janota D, Pang J, Salphati L, Meklemburg R, Korman AJ, Harper HE, Stubblefield S, Payandeh J, McHugh D, Lang BT, Tesar PJ, Dere E, Masureel M, Adams DJ, Volgraf M, Braun MG
J Med Chem (2024) 67 pp. 4819-4832 [ Pubmed: 38470227 DOI: doi:10.1021/acs.jmedchem.3c02396 ]
Structural Basis of Human NOX5 Activation
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8u86
Q-score: 0.45
Cui C, Jiang M, Jain N, Das S, Lo YH, Kermani AA, Pipatpolkai T, Sun J
Nat Commun (2024) 15 pp. 3994-3994 [ DOI: doi:10.1038/s41467-024-48467-y Pubmed: 38734761 ]
- Zinc ion (65 Da, Ligand)
- Nadph oxidase 5 (82 kDa, Protein from Homo sapiens)
- Nadph oxidase 5 (Complex from Homo sapiens)
- Decane (142 Da, Ligand)
- Nadph dihydro-nicotinamide-adenine-dinucleotide phosphate (745 Da, Ligand)
- Heme b/c (618 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Dodecane (170 Da, Ligand)
Structural Basis of Human NOX5 Activation
Resolution: 4.06 Å
EM Method: Single-particle
Fitted PDBs: 8u7y
Q-score: 0.327
Cui C, Jiang M, Jain N, Das S, Lo YH, Kermani AA, Pipatpolkai T, Sun J
Nat Commun (2024) 15 pp. 3994-3994 [ DOI: doi:10.1038/s41467-024-48467-y Pubmed: 38734761 ]
- Zinc ion (65 Da, Ligand)
- Nadph oxidase 5 (82 kDa, Protein from Homo sapiens)
- Nadp nicotinamide-adenine-dinucleotide phosphate (743 Da, Ligand)
- Nadph oxidase 5 (Complex from Homo sapiens)
- Decane (142 Da, Ligand)
- Heme b/c (618 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Dodecane (170 Da, Ligand)
Structure of GABAAR determined by cryoEM at 100 keV
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8pvb
Q-score: 0.486
McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
PNAS (2023) 120 pp. e2312905120-e2312905120 [ Pubmed: 38011573 DOI: doi:10.1073/pnas.2312905120 ]
- Human gaba(a)r-beta3 homopentamer in complex with megabody-25 in lipid nanodisc (Complex from Homo sapiens)
- Histamine (111 Da, Ligand)
- Hexadecane (226 Da, Ligand)
- Decane (142 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit beta-3 (45 kDa, Protein from Homo sapiens)
- Megabody mb25 (56 kDa, Protein from Lama glama)
- Etomidate (244 Da, Ligand)
- Chloride ion (35 Da, Ligand)
structure of a membrane-bound glycosyltransferase
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7xe4
Q-score: 0.471
Hu X, Yang P, Chai C, Liu J, Sun H, Wu Y, Zhang M, Zhang M, Liu X, Yu H
Nature (2023) 616 pp. 190-198 [ Pubmed: 36949198 DOI: doi:10.1038/s41586-023-05856-5 ]
- 1,3-beta-glucan synthase component fks1 (215 kDa, Protein from Saccharomyces cerevisiae)
- Tetradecane (198 Da, Ligand)
- (11r,14s)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate (495 Da, Ligand)
- Decane (142 Da, Ligand)
- Nonane (128 Da, Ligand)
- Heptane (100 Da, Ligand)
- Membrane-bound glycosyltransferase (215 kDa, Complex from Saccharomyces cerevisiae)
- Dodecane (170 Da, Ligand)
Structure of a mutated membrane-bound glycosyltransferase
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7yuy
Q-score: 0.494
Hu X, Yang P, Chai C, Liu J, Sun H, Wu Y, Zhang M, Zhang M, Liu X, Yu H
Nature (2023) 616 pp. 190-198 [ Pubmed: 36949198 DOI: doi:10.1038/s41586-023-05856-5 ]
- Tetradecane (198 Da, Ligand)
- (11r,14s)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate (495 Da, Ligand)
- 1,3-beta-glucan synthase component fks1 (215 kDa, Protein from Saccharomyces cerevisiae)
- Decane (142 Da, Ligand)
- Nonane (128 Da, Ligand)
- Heptane (100 Da, Ligand)
- Membrane-bound glycosyltransferase (215 kDa, Complex from Saccharomyces cerevisiae)
- Dodecane (170 Da, Ligand)
Resolution: 3.03 Å
EM Method: Single-particle
Fitted PDBs: 8b01
Q-score: 0.539
Davies JS, Currie MJ, North RA, Scalise M, Wright JD, Copping JM, Remus DM, Gulati A, Morado DR, Jamieson SA, Newton-Vesty MC, Abeysekera GS, Ramaswamy S, Friemann R, Wakatsuki S, Allison JR, Indiveri C, Drew D, Mace PD, Dobson RCJ
Nat Commun (2023) 14 pp. 1120-1120 [ Pubmed: 36849793 DOI: doi:10.1038/s41467-023-36590-1 ]
- Megabody c7hopq (54 kDa, Protein from synthetic construct)
- Docosane (310 Da, Ligand)
- Hexane (86 Da, Ligand)
- Putative trap-type c4-dicarboxylate transport system, large permease component (45 kDa, Protein from Photobacterium profundum SS9)
- Phosphatidylethanolamine (734 Da, Ligand)
- Putative trap-type c4-dicarboxylate transport system, small permease component (19 kDa, Protein from Photobacterium profundum SS9)
- Sodium ion (22 Da, Ligand)
- Tridecane (184 Da, Ligand)
- Decane (142 Da, Ligand)
- Siaqm (Complex from Photobacterium profundum)
- N-octane (114 Da, Ligand)
- Siaqm with megabody (Complex)
- Megabody (Complex from Helicobacter pylori)
Resolution: 2.79 Å
EM Method: Single-particle
Fitted PDBs: 7pbz
Q-score: 0.548
Kasaragod VB, Mortensen M, Hardwick SW, Wahid AA, Dorovykh V, Chirgadze DY, Smart TG, Miller PS
Nature (2022) 602 pp. 529-533 [ DOI: doi:10.1038/s41586-022-04402-z Pubmed: 35140402 ]
- Gamma-aminobutyric acid receptor subunit alpha-1 (47 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Hexane (86 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit alpha-1 (41 kDa, Protein from Homo sapiens)
- Gaba-a receptor - megabody 25 complex (220 kDa, Complex)
- Undecane (156 Da, Ligand)
- Megabody 25 (Complex from Lama glama)
- Gamma-amino-butanoic acid (103 Da, Ligand)
- Histamine (111 Da, Ligand)
- Decane (142 Da, Ligand)
- N-octane (114 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit beta-3 (51 kDa, Protein from Homo sapiens)
- Gaba-a receptor (Complex from Homo sapiens)
- Megabody 25 (56 kDa, Protein from Lama glama)
Structure of human prestin in the presence of NaCl
Resolution: 2.3 Å
EM Method: Single-particle
Fitted PDBs: 7lgu
Q-score: 0.644
Ge J, Elferich J, Dehghani-Ghahnaviyeh S, Zhao Z, Meadows M, von Gersdorff H, Tajkhorshid E, Gouaux E
Cell (2021) 184 pp. 4669- [ Pubmed: 34390643 DOI: doi:10.1016/j.cell.2021.07.034 ]
- Tetradecane (198 Da, Ligand)
- Hexane (86 Da, Ligand)
- Dodecane (170 Da, Ligand)
- Decane (142 Da, Ligand)
- Prestin (82 kDa, Protein from Homo sapiens)
- Protein structure bound with substrates and lipids (Complex from Homo sapiens)
- N-octane (114 Da, Ligand)
- Heptane (100 Da, Ligand)
- Cholesterol (386 Da, Ligand)
- Water (18 Da, Ligand)
- Chloride ion (35 Da, Ligand)