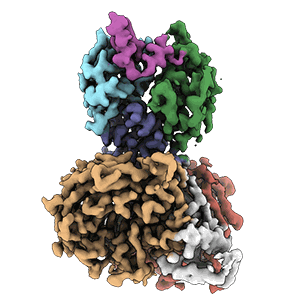
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5
Resolution: 2.63 Å
EM Method: Single-particle
Fitted PDBs: 8tl6
Q-score: 0.598
Radko-Juettner S, Yue H, Myers JA, Carter RD, Robertson AN, Mittal P, Zhu Z, Hansen BS, Donovan KA, Hunkeler M, Rosikiewicz W, Wu Z, McReynolds MG, Roy Burman SS, Schmoker AM, Mageed N, Brown SA, Mobley RJ, Partridge JF, Stewart EA, Pruett-Miller SM, Nabet B, Peng J, Gray NS, Fischer ES, Roberts CWM
Nature (2024) 628 pp. 442-449 [ Pubmed: 38538798 DOI: doi:10.1038/s41586-024-07250-1 ]
- Det1- and ddb1-associated protein 1 (14 kDa, Protein from Homo sapiens)
- Complex of ddb1deltab-dda1-dcaf5 (220 kDa, Complex from Homo sapiens)
- Dna damage-binding protein 1 (96 kDa, Protein from Homo sapiens)
- Ddb1- and cul4-associated factor 5 (108 kDa, Protein from Homo sapiens)
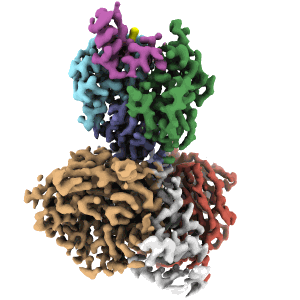
Cryo-EM structure of DDB1dB:CRBN:Pomalidomide:SD40
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8tnp
Q-score: 0.507
Mercer JAM, DeCarlo SJ, Roy Burman SS, Sreekanth V, Nelson AT, Hunkeler M, Chen PJ, Donovan KA, Kokkonda P, Tiwari PK, Shoba VM, Deb A, Choudhary A, Fischer ES, Liu DR
Science (2024) 383 pp. eadk4422-eadk4422 [ DOI: doi:10.1126/science.adk4422 Pubmed: 38484051 ]
- Protein cereblon (55 kDa, Protein from Homo sapiens)
- S-pomalidomide (273 Da, Ligand)
- Dna damage-binding protein 1 (96 kDa, Protein from Homo sapiens)
- Ternary complex of ddb1db:crbn:pomalidomide with sd40 (201 kDa, Complex)
- Zinc ion (65 Da, Ligand)
- Maltose/maltodextrin-binding periplasmic protein,sd40 (50 kDa, Protein from Homo sapiens)
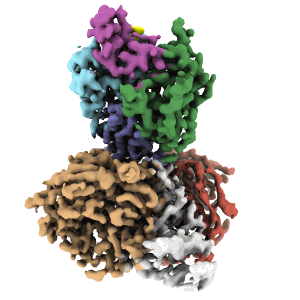
Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 1
Resolution: 2.41 Å
EM Method: Single-particle
Fitted PDBs: 8tnq
Q-score: 0.591
Mercer JAM, DeCarlo SJ, Roy Burman SS, Sreekanth V, Nelson AT, Hunkeler M, Chen PJ, Donovan KA, Kokkonda P, Tiwari PK, Shoba VM, Deb A, Choudhary A, Fischer ES, Liu DR
Science (2024) 383 pp. eadk4422-eadk4422 [ DOI: doi:10.1126/science.adk4422 Pubmed: 38484051 ]
- Protein cereblon (55 kDa, Protein from Homo sapiens)
- 2-[(3s)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1h-isoindole-1,3(2h)-dione (343 Da, Ligand)
- Ternary complex of ddb1db:crbn:pt-179 with sd40 (201 kDa, Complex)
- Dna damage-binding protein 1 (96 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Maltose/maltodextrin-binding periplasmic protein,sd40 (50 kDa, Protein from Homo sapiens)
Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8tnr
Q-score: 0.561
Mercer JAM, DeCarlo SJ, Roy Burman SS, Sreekanth V, Nelson AT, Hunkeler M, Chen PJ, Donovan KA, Kokkonda P, Tiwari PK, Shoba VM, Deb A, Choudhary A, Fischer ES, Liu DR
Science (2024) 383 pp. eadk4422-eadk4422 [ Pubmed: 38484051 DOI: doi:10.1126/science.adk4422 ]
- Ternary complex of ddb1db:crbn:pt-179 with sd40 (201 kDa, Complex)
- 2-[(3s)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1h-isoindole-1,3(2h)-dione (343 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna damage-binding protein 1 (96 kDa, Protein from Homo sapiens)
- Maltose/maltodextrin-binding periplasmic protein,sd40 (50 kDa, Protein from Homo sapiens)
- Protein cereblon (55 kDa, Protein from Homo sapiens)
Map from local refinement (focused on CRBN) of DDB1dB:CRBN:Pomalidomide:SD40
Mercer JAM, DeCarlo SJ, Roy Burman SS, Sreekanth V, Nelson AT, Hunkeler M, Chen PJ, Donovan KA, Kokkonda P, Tiwari PK, Shoba VM, Deb A, Choudhary A, Fischer ES, Liu DR
Science (2024) 383 pp. eadk4422-eadk4422 [ DOI: doi:10.1126/science.adk4422 Pubmed: 38484051 ]
Map from local refinement (focused on CRBN) of DDB1dB:CRBN:PT-179:SD40, conformation 1
Mercer JAM, DeCarlo SJ, Roy Burman SS, Sreekanth V, Nelson AT, Hunkeler M, Chen PJ, Donovan KA, Kokkonda P, Tiwari PK, Shoba VM, Deb A, Choudhary A, Fischer ES, Liu DR
Science (2024) 383 pp. eadk4422-eadk4422 [ DOI: doi:10.1126/science.adk4422 Pubmed: 38484051 ]
Map from local refinement (focused on CRBN) of DDB1dB:CRBN:PT-179:SD40, conformation 2
Mercer JAM, DeCarlo SJ, Roy Burman SS, Sreekanth V, Nelson AT, Hunkeler M, Chen PJ, Donovan KA, Kokkonda P, Tiwari PK, Shoba VM, Deb A, Choudhary A, Fischer ES, Liu DR
Science (2024) 383 pp. eadk4422-eadk4422 [ DOI: doi:10.1126/science.adk4422 Pubmed: 38484051 ]
Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2
Resolution: 2.2 Å
EM Method: Single-particle
Fitted PDBs: 8g46
Q-score: 0.643
Li YD, Ma MW, Hassan MM, Hunkeler M, Teng M, Puvar K, Lumpkin R, Sandoval B, Jin CY, Ficarro SB, Wang MY, Xu S, Groendyke BJ, Sigua LH, Tavares I, Zou C, Tsai JM, Park PMC, Yoon H, Majewski FC, Marto JA, Qi J, Nowak RP, Donovan KA, Slabicki M, Gray NS, Fischer ES, Ebert BL
bioRxiv (2023) [ DOI: doi:10.1101/2023.02.14.528208 Pubmed: 36824856 ]
- Water (18 Da, Ligand)
- Ddb1- and cul4-associated factor 16 (24 kDa, Protein from Homo sapiens)
- Tert-butyl [(6s,10p)-4-{4-[(ethanesulfonyl)amino]phenyl}-2,3,9-trimethyl-6h-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate (529 Da, Ligand)
- Bromodomain-containing protein 4 (14 kDa, Protein from Homo sapiens)
- Ternary complex of cullin ring e3 ubiquitin ligase substrate receptor arm (ddb1deltab-dda1-dcaf16) in compound-induced complex with bromodomain 2 of brd4 (148 kDa, Complex from Homo sapiens)
- Det1- and ddb1-associated protein 1 (12 kDa, Protein from Homo sapiens)
- Dna damage-binding protein 1 (96 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)