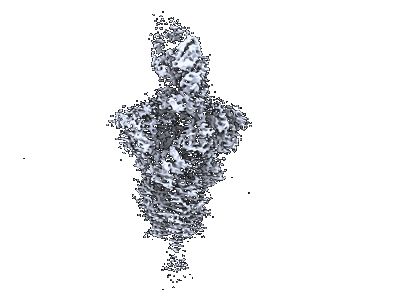Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 9asd
Q-score: 0.506
Rosen LE, Tortorici MA, De Marco A, Pinto D, Foreman WB, Taylor AL, Park YJ, Bohan D, Rietz T, Errico JM, Hauser K, Dang HV, Chartron JW, Giurdanella M, Cusumano G, Saliba C, Zatta F, Sprouse KR, Addetia A, Zepeda SK, Brown J, Lee J, Dellota Jr E, Rajesh A, Noack J, Tao Q, DaCosta Y, Tsu B, Acosta R, Subramanian S, de Melo GD, Kergoat L, Zhang I, Liu Z, Guarino B, Schmid MA, Schnell G, Miller JL, Lempp FA, Czudnochowski N, Cameroni E, Whelan SPJ, Bourhy H, Purcell LA, Benigni F, di Iulio J, Pizzuto MS, Lanzavecchia A, Telenti A, Snell G, Corti D, Veesler D, Starr TN
Cell (2024) 187 pp. 7196- [ DOI: doi:10.1016/j.cell.2024.09.026 Pubmed: 39383863 ]
- Vir-7229 fab light chain (11 kDa, Protein from Homo sapiens)
- Spike glycoprotein (141 kDa, Protein from Homo sapiens)
- Vir-7229 fab heavy chain (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ba 2.86 rbd - s2v29 complex (local refinement) (Complex from Sarbecovirus)
VIR-7229 Fab fragment bound the BA.2.86 spike trimer (global refinement)
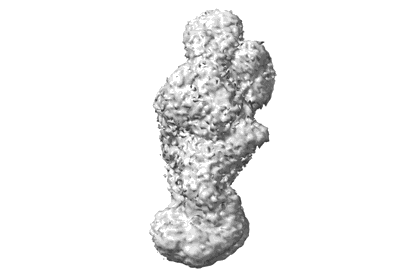
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 9au2
Q-score: 0.499
Rosen LE, Tortorici MA, De Marco A, Pinto D, Foreman WB, Taylor AL, Park YJ, Bohan D, Rietz T, Errico JM, Hauser K, Dang HV, Chartron JW, Giurdanella M, Cusumano G, Saliba C, Zatta F, Sprouse KR, Addetia A, Zepeda SK, Brown J, Lee J, Dellota Jr E, Rajesh A, Noack J, Tao Q, DaCosta Y, Tsu B, Acosta R, Subramanian S, de Melo GD, Kergoat L, Zhang I, Liu Z, Guarino B, Schmid MA, Schnell G, Miller JL, Lempp FA, Czudnochowski N, Cameroni E, Whelan SPJ, Bourhy H, Purcell LA, Benigni F, di Iulio J, Pizzuto MS, Lanzavecchia A, Telenti A, Snell G, Corti D, Veesler D, Starr TN
Cell (2024) 187 pp. 7196- [ DOI: doi:10.1016/j.cell.2024.09.026 Pubmed: 39383863 ]
- Vir-7229 fab light chain (23 kDa, Protein from Homo sapiens)
- Vir-7229 fab heavy chain (24 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ba.2.86 spike glycoprotein in complex with vir-7229 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab
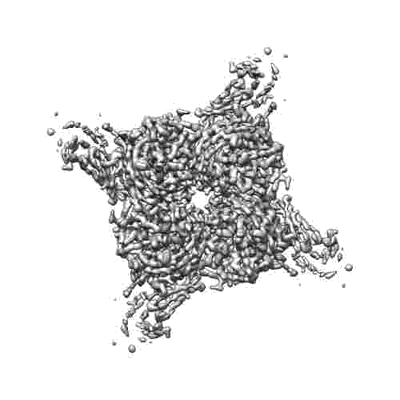
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8qtd
Q-score: 0.454
Liu C, Zhou D, Dijokaite-Guraliuc A, Supasa P, Duyvesteyn HME, Ginn HM, Selvaraj M, Mentzer AJ, Das R, de Silva TI, Ritter TG, Plowright M, Newman TAH, Stafford L, Kronsteiner B, Temperton N, Lui Y, Fellermeyer M, Goulder P, Klenerman P, Dunachie SJ, Barton MI, Kutuzov MA, Dushek O, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR
Cell Rep Med (2024) 5 pp. 101553-101553 [ Pubmed: 38723626 DOI: doi:10.1016/j.xcrm.2024.101553 ]
- Spike glycoprotein,fibritin (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Xbb-7 fab light chain (10 kDa, Protein from Homo sapiens)
- Xbb-7 fab heavy chain (14 kDa, Protein from Homo sapiens)
- Ba-2.86 variant sars-cov2 s protein complexed with xbb-7 fab. (Complex)
- Ba.2.86 variant sars-cov2 s protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Xbb-7 fab (Complex from Homo sapiens)
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
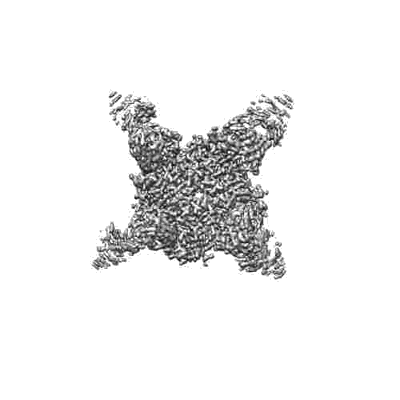
Resolution: 3.41 Å
EM Method: Single-particle
Fitted PDBs: 8r8k
Q-score: 0.504
Liu C, Zhou D, Dijokaite-Guraliuc A, Supasa P, Duyvesteyn HME, Ginn HM, Selvaraj M, Mentzer AJ, Das R, de Silva TI, Ritter TG, Plowright M, Newman TAH, Stafford L, Kronsteiner B, Temperton N, Lui Y, Fellermeyer M, Goulder P, Klenerman P, Dunachie SJ, Barton MI, Kutuzov MA, Dushek O, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR
Cell Rep Med (2024) 5 pp. 101553-101553 [ Pubmed: 38723626 DOI: doi:10.1016/j.xcrm.2024.101553 ]
- Ba.2.12.1 spike glycoprotein ectodomain (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein,fibritin (142 kDa, Protein from Tequatrovirus T4)
- Xbb-4 fab heavy chain (24 kDa, Protein from Homo sapiens)
- Xbb-4 fab in complex with sars-cov-2 ba.2.12.1 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Xbb-4 fab (Complex from Homo sapiens)
- Xbb-4 fab light chain (23 kDa, Protein from Homo sapiens)
Structure of KRAS G12V/HLA-A*03:01 in complex with antibody fragment V2
Resolution: 3.14 Å
EM Method: Single-particle
Fitted PDBs: 7stf
Q-score: 0.393
Wright KM, DiNapoli SR, Miller MS, Aitana Azurmendi P, Zhao X, Yu Z, Chakrabarti M, Shi W, Douglass J, Hwang MS, Hsiue EH, Mog BJ, Pearlman AH, Paul S, Konig MF, Pardoll DM, Bettegowda C, Papadopoulos N, Kinzler KW, Vogelstein B, Zhou S, Gabelli SB
Nat Commun (2023) 14 pp. 5063-5063 [ Pubmed: 37604828 DOI: doi:10.1038/s41467-023-40821-w ]
- Ternary complex of antibody-fragment v2 with kras g12v/hla-a*03:01 (Complex)
- Igg, fab heavy chain v2 (23 kDa, Protein from Homo sapiens)
- Kras g12v (7-16) (885 Da, Protein from synthetic construct)
- Hla class i histocompatibility antigen, a alpha chain (32 kDa, Protein from Homo sapiens)
- Igg, fab light chain v2 (23 kDa, Protein from Homo sapiens)
- Beta-2-microglobulin (11 kDa, Protein from Homo sapiens)
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 4 FNI19 Fab molecules
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8g30
Q-score: 0.487
Momont C, Dang HV, Zatta F, Hauser K, Wang C, di Iulio J, Minola A, Czudnochowski N, De Marco A, Branch K, Donermeyer D, Vyas S, Chen A, Ferri E, Guarino B, Powell AE, Spreafico R, Yim SS, Balce DR, Bartha I, Meury M, Croll TI, Belnap DM, Schmid MA, Schaiff WT, Miller JL, Cameroni E, Telenti A, Virgin HW, Rosen LE, Purcell LA, Lanzavecchia A, Snell G, Corti D, Pizzuto MS
Nature (2023) 618 pp. 590-597 [ Pubmed: 37258672 DOI: doi:10.1038/s41586-023-06136-y ]
- Fni19 fab heavy chain (24 kDa, Protein from Homo sapiens)
- N2 neuraminidase of a/tanzania/205/2010 h3n2 in complex with 4 fni19 fab molecules (420 kDa, Complex from Influenza A virus)
- Neuraminidase (54 kDa, Protein from Influenza A virus)
- Calcium ion (40 Da, Ligand)
- Fni19 fab light chain (23 kDa, Protein from Homo sapiens)
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 3 FNI9 Fab molecules
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8g3m
Q-score: 0.502
Momont C, Dang HV, Zatta F, Hauser K, Wang C, di Iulio J, Minola A, Czudnochowski N, De Marco A, Branch K, Donermeyer D, Vyas S, Chen A, Ferri E, Guarino B, Powell AE, Spreafico R, Yim SS, Balce DR, Bartha I, Meury M, Croll TI, Belnap DM, Schmid MA, Schaiff WT, Miller JL, Cameroni E, Telenti A, Virgin HW, Rosen LE, Purcell LA, Lanzavecchia A, Snell G, Corti D, Pizzuto MS
Nature (2023) 618 pp. 590-597 [ Pubmed: 37258672 DOI: doi:10.1038/s41586-023-06136-y ]
- Fni9 fab light chain (23 kDa, Protein from Homo sapiens)
- Fni9 fab heavy chain (24 kDa, Protein from Homo sapiens)
- Neuraminidase (54 kDa, Protein from Influenza A virus)
- N2 neuraminidase of a/tanzania/205/2010 h3n2 in complex with 3 fni9 fab molecules (420 kDa, Complex from Influenza A virus)
- Calcium ion (40 Da, Ligand)
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 4 FNI9 Fab molecules
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8g3n
Q-score: 0.549
Momont C, Dang HV, Zatta F, Hauser K, Wang C, di Iulio J, Minola A, Czudnochowski N, De Marco A, Branch K, Donermeyer D, Vyas S, Chen A, Ferri E, Guarino B, Powell AE, Spreafico R, Yim SS, Balce DR, Bartha I, Meury M, Croll TI, Belnap DM, Schmid MA, Schaiff WT, Miller JL, Cameroni E, Telenti A, Virgin HW, Rosen LE, Purcell LA, Lanzavecchia A, Snell G, Corti D, Pizzuto MS
Nature (2023) 618 pp. 590-597 [ Pubmed: 37258672 DOI: doi:10.1038/s41586-023-06136-y ]
- Fni9 fab light chain (23 kDa, Protein from Homo sapiens)
- Fni9 fab heavy chain (24 kDa, Protein from Homo sapiens)
- 4 fni9 fabs in complex with tetrameric n2 na protein from a/tanzania/2010 strain (420 kDa, Complex from Influenza A virus)
- Neuraminidase (54 kDa, Protein from Influenza A virus)
- Calcium ion (40 Da, Ligand)
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI9 Fab molecules
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8g3o
Q-score: 0.517
Momont C, Dang HV, Zatta F, Hauser K, Wang C, di Iulio J, Minola A, Czudnochowski N, De Marco A, Branch K, Donermeyer D, Vyas S, Chen A, Ferri E, Guarino B, Powell AE, Spreafico R, Yim SS, Balce DR, Bartha I, Meury M, Croll TI, Belnap DM, Schmid MA, Schaiff WT, Miller JL, Cameroni E, Telenti A, Virgin HW, Rosen LE, Purcell LA, Lanzavecchia A, Snell G, Corti D, Pizzuto MS
Nature (2023) 618 pp. 590-597 [ Pubmed: 37258672 DOI: doi:10.1038/s41586-023-06136-y ]
- Fni9 fab light chain (23 kDa, Protein from Homo sapiens)
- 3 fni9 fabs in complex with tetrameric n2 na protein from a/hong_kong/2671/2019 strain (420 kDa, Complex from Influenza A virus)
- Neuraminidase (54 kDa, Protein from Influenza A virus)
- Fni9 fab heavy chain (24 kDa, Protein from Homo sapiens)
- Calcium ion (40 Da, Ligand)
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 4 FNI9 Fab molecules
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8g3p
Q-score: 0.588
Momont C, Dang HV, Zatta F, Hauser K, Wang C, di Iulio J, Minola A, Czudnochowski N, De Marco A, Branch K, Donermeyer D, Vyas S, Chen A, Ferri E, Guarino B, Powell AE, Spreafico R, Yim SS, Balce DR, Bartha I, Meury M, Croll TI, Belnap DM, Schmid MA, Schaiff WT, Miller JL, Cameroni E, Telenti A, Virgin HW, Rosen LE, Purcell LA, Lanzavecchia A, Snell G, Corti D, Pizzuto MS
Nature (2023) 618 pp. 590-597 [ Pubmed: 37258672 DOI: doi:10.1038/s41586-023-06136-y ]
- Fni9 fab light chain (23 kDa, Protein from Homo sapiens)
- Neuraminidase (54 kDa, Protein from Influenza A virus)
- Fni9 fab heavy chain (24 kDa, Protein from Homo sapiens)
- N2 neuraminidase of a/hong_kong/2671/2019 in complex with 4 fni9 fab molecules (420 kDa, Complex from Influenza A virus)
- Calcium ion (40 Da, Ligand)