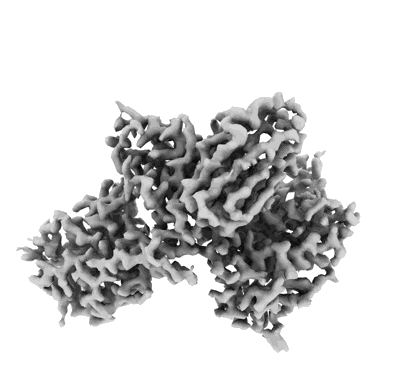
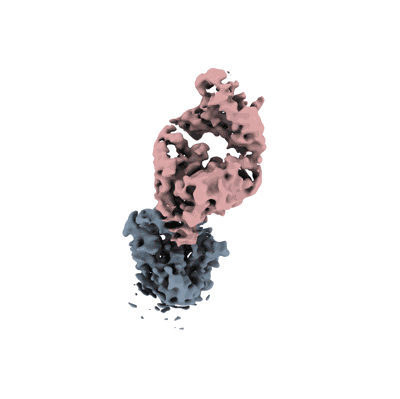Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 9asd
Q-score: 0.506
Rosen LE, Tortorici MA, De Marco A, Pinto D, Foreman WB, Taylor AL, Park YJ, Bohan D, Rietz T, Errico JM, Hauser K, Dang HV, Chartron JW, Giurdanella M, Cusumano G, Saliba C, Zatta F, Sprouse KR, Addetia A, Zepeda SK, Brown J, Lee J, Dellota Jr E, Rajesh A, Noack J, Tao Q, DaCosta Y, Tsu B, Acosta R, Subramanian S, de Melo GD, Kergoat L, Zhang I, Liu Z, Guarino B, Schmid MA, Schnell G, Miller JL, Lempp FA, Czudnochowski N, Cameroni E, Whelan SPJ, Bourhy H, Purcell LA, Benigni F, di Iulio J, Pizzuto MS, Lanzavecchia A, Telenti A, Snell G, Corti D, Veesler D, Starr TN
Cell (2024) 187 pp. 7196- [ DOI: doi:10.1016/j.cell.2024.09.026 Pubmed: 39383863 ]
- Vir-7229 fab light chain (11 kDa, Protein from Homo sapiens)
- Spike glycoprotein (141 kDa, Protein from Homo sapiens)
- Vir-7229 fab heavy chain (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ba 2.86 rbd - s2v29 complex (local refinement) (Complex from Sarbecovirus)
VIR-7229 Fab fragment bound the BA.2.86 spike trimer (global refinement)
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 9au2
Q-score: 0.499
Rosen LE, Tortorici MA, De Marco A, Pinto D, Foreman WB, Taylor AL, Park YJ, Bohan D, Rietz T, Errico JM, Hauser K, Dang HV, Chartron JW, Giurdanella M, Cusumano G, Saliba C, Zatta F, Sprouse KR, Addetia A, Zepeda SK, Brown J, Lee J, Dellota Jr E, Rajesh A, Noack J, Tao Q, DaCosta Y, Tsu B, Acosta R, Subramanian S, de Melo GD, Kergoat L, Zhang I, Liu Z, Guarino B, Schmid MA, Schnell G, Miller JL, Lempp FA, Czudnochowski N, Cameroni E, Whelan SPJ, Bourhy H, Purcell LA, Benigni F, di Iulio J, Pizzuto MS, Lanzavecchia A, Telenti A, Snell G, Corti D, Veesler D, Starr TN
Cell (2024) 187 pp. 7196- [ DOI: doi:10.1016/j.cell.2024.09.026 Pubmed: 39383863 ]
- Vir-7229 fab light chain (23 kDa, Protein from Homo sapiens)
- Vir-7229 fab heavy chain (24 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ba.2.86 spike glycoprotein in complex with vir-7229 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
the complex structure of the H4B6 Fab with the RBD of Omicron BA.5 S protein
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8ywx
Q-score: 0.387
Sun H, Xia L, Li J, Zhang Y, Zhang G, Huang P, Wang X, Cui Y, Fang T, Fan P, Zhou Q, Chi X, Yu C
Emerg Microbes Infect (2024) 13 pp. 2373307-2373307 [ DOI: doi:10.1080/22221751.2024.2373307 Pubmed: 38953857 ]
- The complex structure of the h4b6 fab with the rbd of omicron ba.5 s protein (Complex from Homo sapiens)
- Spike protein s2' (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- H4b6 fab's heavy chain (76 kDa, Protein from Homo sapiens)
- H4b6 fab's light chain (23 kDa, Protein from Homo sapiens)
SARS-CoV-2 Spike S2 bound to Fab 54043-5
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8vcr
Q-score: 0.535
Johnson NV, Wall SC, Kramer KJ, Holt CM, Periasamy S, Richardson SI, Manamela NP, Suryadevara N, Andreano E, Paciello I, Pierleoni G, Piccini G, Huang Y, Ge P, Allen JD, Uno N, Shiakolas AR, Pilewski KA, Nargi RS, Sutton RE, Abu-Shmais AA, Parks R, Haynes BF, Carnahan RH, Crowe Jr JE, Montomoli E, Rappuoli R, Bukreyev A, Ross TM, Sautto GA, McLellan JS, Georgiev IS
Structure (2024) 32 pp. 1893- [ Pubmed: 39326419 DOI: doi:10.1016/j.str.2024.08.022 ]
- S2 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike protein s2 (65 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab 54043-5 (Complex from Homo sapiens)
- 54043-5 fab light chain (23 kDa, Protein from Homo sapiens)
- Trimeric s2 bound to fab 54043-5 (Complex)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 54043-5 fab heavy chain (25 kDa, Protein from Homo sapiens)
SARS-CoV-2 spike omicron (BA.1) ectodomain trimer in complex with SC27 Fab, local refinement
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8vif
Q-score: 0.545
Voss WN, Mallory MA, Byrne PO, Marchioni JM, Knudson SA, Powers JM, Leist SR, Dadonaite B, Townsend DR, Kain J, Huang Y, Satterwhite E, Castillo IN, Mattocks M, Paresi C, Munt JE, Scobey T, Seeger A, Premkumar L, Bloom JD, Georgiou G, McLellan JS, Baric RS, Lavinder JJ, Ippolito GC
Cell Rep Med (2024) 5 pp. 101668-101668 [ DOI: doi:10.1016/j.xcrm.2024.101668 Pubmed: 39094579 ]
- Sc27 fab light chain (11 kDa, Protein from Homo sapiens)
- Sc27 fab heavy chain (14 kDa, Protein from Homo sapiens)
- Sars-cov-2 spike omicron (ba.1) ectodomain trimer in complex with sc27 fab, global refinement (Complex)
- Sars-cov-2 spike omicron (ba.1) ectodomain (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (144 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sc27 fab (Complex from Homo sapiens)
SARS-CoV-2 spike omicron (BA.1) RBD ectodomain dimer-of-trimers in complex with SC27 Fabs
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8vke
Q-score: 0.544
Voss WN, Mallory MA, Byrne PO, Marchioni JM, Knudson SA, Powers JM, Leist SR, Dadonaite B, Townsend DR, Kain J, Huang Y, Satterwhite E, Castillo IN, Mattocks M, Paresi C, Munt JE, Scobey T, Seeger A, Premkumar L, Bloom JD, Georgiou G, McLellan JS, Baric RS, Lavinder JJ, Ippolito GC
Cell Rep Med (2024) 5 pp. 101668-101668 [ DOI: doi:10.1016/j.xcrm.2024.101668 Pubmed: 39094579 ]
- Sars-cov-2 spike omicron (ba.1) rbd ectodomain dimer-of-trimers in complex with sc27 fabs (Complex)
- Sc27 fab light chain (25 kDa, Protein from Homo sapiens)
- Spike glycoprotein (144 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sc27 fab heavy chain (27 kDa, Protein from Homo sapiens)
Complex of NPR1 ectodomain with ANP plus an allosteric activating antibody, REGN5381
Resolution: 3.08 Å
EM Method: Single-particle
Fitted PDBs: 8tg9
Q-score: 0.495
Dunn ME, Kithcart A, Kim JH, Ho AJ, Franklin MC, Romero Hernandez A, de Hoon J, Botermans W, Meyer J, Jin X, Zhang D, Torello J, Jasewicz D, Kamat V, Garnova E, Liu N, Rosconi M, Pan H, Karnik S, Burczynski ME, Zheng W, Rafique A, Nielsen JB, De T, Verweij N, Pandit A, Locke A, Chalasani N, Melander O, Schwantes-An TH, Baras A, Lotta LA, Musser BJ, Mastaitis J, Devalaraja-Narashimha KB, Rankin AJ, Huang T, Herman G, Olson W, Murphy AJ, Yancopoulos GD, Olenchock BA, Morton L
Nature (2024) 633 pp. 654-661 [ DOI: doi:10.1038/s41586-024-07903-1 Pubmed: 39261724 ]
- Regn5381 fab heavy chain (24 kDa, Protein from Mus musculus)
- Atrial natriuretic peptide receptor 1 (52 kDa, Protein from Homo sapiens)
- Regn5381 fab light chain (23 kDa, Protein from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Active complex of npr1 ectodomain, anp, and regn5381 fab (Complex from Homo sapiens)
- Atrial natriuretic peptide (3 kDa, Protein from Homo sapiens)
Cryo-EM structure of bsAb3 Fab-Gn-Gc complex
Resolution: 3.27 Å
EM Method: Single-particle
Fitted PDBs: 8wqw
Q-score: 0.349
Chang Z, Gao D, Liao L, Sun J, Zhang G, Zhang X, Wang F, Li C, Oladejo BO, Li S, Chai Y, Hu Y, Lu X, Xiao H, Qi J, Chen Z, Gao F, Wu Y
PNAS (2024) 121 pp. e2400163121-e2400163121 [ Pubmed: 38830098 DOI: doi:10.1073/pnas.2400163121 ]
- Glycoprotein n (35 kDa, Protein from Dabie bandavirus)
- Glycoprotein c (47 kDa, Protein from Dabie bandavirus)
- Gn_gc (Complex from Dabie bandavirus)
- Fab (Complex from Homo sapiens)
- Bsab3 fab-heavy chain (37 kDa, Protein from Homo sapiens)
- Bsab3 fab-light chain (35 kDa, Protein from Homo sapiens)
- The structure of bsab3_gn_gc complex (Complex)
Local refinement of SARS-CoV-2 (HP-GSAS-Mut7) spike NTD in complex with TXG-0078 Fab
Resolution: 3.88 Å
EM Method: Single-particle
Fitted PDBs: 8swh
Q-score: 0.326
Hurtado J, Rogers TF, Jaffe DB, Adams BA, Bangaru S, Garcia E, Capozzola T, Messmer T, Sharma P, Song G, Beutler N, He W, Dueker K, Musharrafieh R, Burbach S, Truong A, Stubbington MJT, Burton DR, Andrabi R, Ward AB, McDonnell WJ, Briney B
Cell Rep (2024) 43 pp. 114307-114307 [ DOI: doi:10.1016/j.celrep.2024.114307 Pubmed: 38848216 ]
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Structure of sars-cov-2(hp-gsas-mut7) spike in complex with txg-0078 fab- local refinement (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Txg-0078 fab heavy chain (16 kDa, Protein from Homo sapiens)
- Txg-0078 fab light chain (14 kDa, Protein from Homo sapiens)
Antibody N3-1 bound to RBDs in the up and down conformations
Resolution: 2.79 Å
EM Method: Single-particle
Fitted PDBs: 8tm1
Q-score: 0.543
Goike J, Hsieh CL, Horton AP, Gardner EC, Zhou L, Bartzoka F, Wang N, Javanmardi K, Herbert A, Abbassi S, Xie X, Xia H, Shi PY, Renberg R, Segall-Shapiro TH, Terrace CI, Wu W, Shroff R, Byrom M, Ellington AD, Marcotte EM, Musser JM, Kuchipudi SV, Kapur V, Georgiou G, Weaver SC, Dye JM, Boutz DR, McLellan JS, Gollihar JD
Commun Biol (2023) 6 pp. 1250-1250 [ Pubmed: 38082099 DOI: doi:10.1038/s42003-023-05649-6 ]
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- N3-1 fab heavy chain (14 kDa, Protein from Homo sapiens)
- Spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- N3-1 fab light chain (11 kDa, Protein from Homo sapiens)
- N3-1 fab (Complex from Homo sapiens)
- The complex of n3-1 fab bound to two receptor binding domains of the spike (Complex)