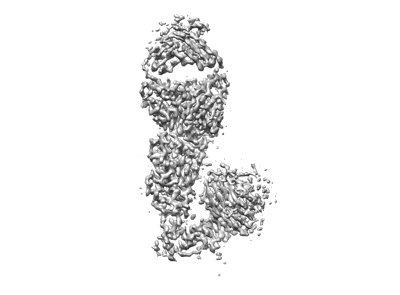
sd1.040 Fab in complex with SARS-CoV-2 Spike 2P glycoprotein
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 8d48
Q-score: 0.449
Bianchini F, Crivelli V, Abernathy ME, Guerra C, Palus M, Muri J, Marcotte H, Piralla A, Pedotti M, De Gasparo R, Simonelli L, Matkovic M, Toscano C, Biggiogero M, Calvaruso V, Svoboda P, Cervantes Rincon T, Fava T, Podesvova L, Shanbhag AA, Celoria A, Sgrignani J, Stefanik M, Honig V, Pranclova V, Michalcikova T, Prochazka J, Guerrini G, Mehn D, Ciabattini A, Abolhassani H, Jarrossay D, Uguccioni M, Medaglini D, Pan-Hammarstrom Q, Calzolai L, Fernandez D, Baldanti F, Franzetti-Pellanda A, Garzoni C, Sedlacek R, Ruzek D, Varani L, Cavalli A, Barnes CO, Robbiani DF
Sci Immunol (2023) 8 pp. eade0958-eade0958 [ Pubmed: 36701425 DOI: doi:10.1126/sciimmunol.ade0958 ]
- Sd1.040 fab heavy chain (24 kDa, Protein from Homo sapiens)
- Spike glycoprotein (139 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sd1.040 fab heavy chain, sd1.040 fab light chain (Complex from Homo sapiens)
- Spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sd1.040 fab light chain (24 kDa, Protein from Homo sapiens)
- Sd1.040 fab in complex with sars-cov-2 spike 2p (79 kDa, Complex)
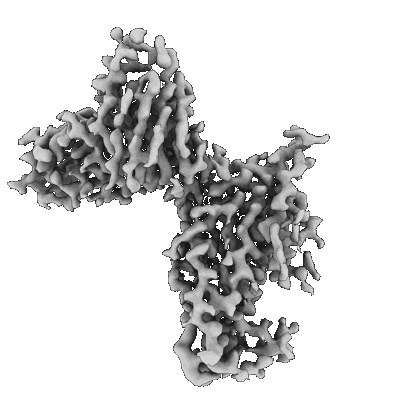
SARS-CoV-2 spike trimer in complex with Fab NE12, local refinement map
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7u9o
Q-score: 0.518
Chen Z, Zhang P, Matsuoka Y, Tsybovsky Y, West K, Santos C, Boyd LF, Nguyen H, Pomerenke A, Stephens T, Olia AS, Zhang B, De Giorgi V, Holbrook MR, Gross R, Postnikova E, Garza NL, Johnson RF, Margulies DH, Kwong PD, Alter HJ, Buchholz UJ, Lusso P, Farci P
Cell Rep (2022) 41 pp. 111528-111528 [ Pubmed: 36302375 DOI: doi:10.1016/j.celrep.2022.111528 ]
- Ne12 fab light chain (23 kDa, Protein from Homo sapiens)
- Sars-cov-2 spike receptor-binding domain in complex with fab ne12 (110 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Ne12 fab heavy chain (24 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 spike trimer in complex with Fab NA8, local refinement map
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7u9p
Q-score: 0.528
Chen Z, Zhang P, Matsuoka Y, Tsybovsky Y, West K, Santos C, Boyd LF, Nguyen H, Pomerenke A, Stephens T, Olia AS, Zhang B, De Giorgi V, Holbrook MR, Gross R, Postnikova E, Garza NL, Johnson RF, Margulies DH, Kwong PD, Alter HJ, Buchholz UJ, Lusso P, Farci P
Cell Rep (2022) 41 pp. 111528-111528 [ Pubmed: 36302375 DOI: doi:10.1016/j.celrep.2022.111528 ]
- Sars-cov-2 spike receptor-binding domain in complex with fab na8 (110 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Na8 fab light chain (23 kDa, Protein from Homo sapiens)
- Na8 fab heavy chain (24 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
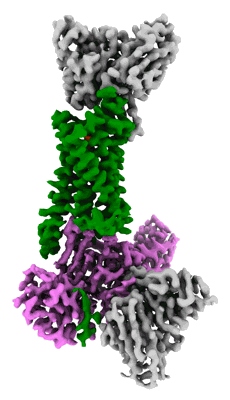
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8erq
Q-score: 0.493
Park YJ, Pinto D, Walls AC, Liu Z, De Marco A, Benigni F, Zatta F, Silacci-Fregni C, Bassi J, Sprouse KR, Addetia A, Bowen JE, Stewart C, Giurdanella M, Saliba C, Guarino B, Schmid MA, Franko NM, Logue JK, Dang HV, Hauser K, di Iulio J, Rivera W, Schnell G, Rajesh A, Zhou J, Farhat N, Kaiser H, Montiel-Ruiz M, Noack J, Lempp FA, Janer J, Abdelnabi R, Maes P, Ferrari P, Ceschi A, Giannini O, de Melo GD, Kergoat L, Bourhy H, Neyts J, Soriaga L, Purcell LA, Snell G, Whelan SPJ, Lanzavecchia A, Virgin HW, Piccoli L, Chu HY, Pizzuto MS, Corti D, Veesler D
Science (2022) 378 pp. 619-627 [ Pubmed: 36264829 DOI: doi:10.1126/science.adc9127 ]
- S2x324 fab heavy chain (13 kDa, Protein from Homo sapiens)
- S2x324 fab light chain (11 kDa, Protein from Homo sapiens)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 ba.1 spike ectodomain trimer in complex with the s2x324 neutralizing antibody fab fragment (local refinement of the rbd and s2x324) (Complex from Severe acute respiratory syndrome coronavirus 2)
- S2x324 fab (Complex from Homo sapiens)
- Sars-cov-2 omicron ba.1 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
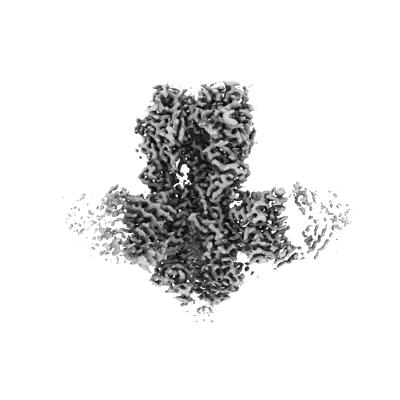
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7srs
Q-score: 0.523
Cao C, Barros-Alvarez X, Zhang S, Kim K, Damgen MA, Panova O, Suomivuori CM, Fay JF, Zhong X, Krumm BE, Gumpper RH, Seven AB, Robertson MJ, Krogan NJ, Huttenhain R, Nichols DE, Dror RO, Skiniotis G, Roth BL
Neuron (2022) 110 pp. 3154- [ Pubmed: 36087581 DOI: doi:10.1016/j.neuron.2022.08.006 ]
- Anti-5ht2br fab heavy chain (23 kDa, Protein from Mus musculus)
- Fab30 light chain (11 kDa, Protein from Mus musculus)
- Isoform 1b of beta-arrestin-1 (41 kDa, Protein from Homo sapiens)
- 5-ht2b receptor bound to lsd in complex with beta-arrestin1 (Complex)
- Anti-5ht2br fab light chain (22 kDa, Protein from Mus musculus)
- Fab30 heavy chain (13 kDa, Protein from Mus musculus)
- (8alpha)-n,n-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide (323 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 5-hydroxytryptamine receptor 2b (37 kDa, Protein from Homo sapiens)
Structure of H1 NC99 influenza hemagglutinin bound to Fab 310-63E6
Resolution: 3.02 Å
EM Method: Single-particle
Fitted PDBs: 7scn
Q-score: 0.534
Sangesland M, Torrents de la Pena A, Boyoglu-Barnum S, Ronsard L, Mohamed FAN, Moreno TB, Barnes RM, Rohrer D, Lonberg N, Ghebremichael M, Kanekiyo M, Ward A, Lingwood D
Immunity (2022) 55 pp. 1693-1709.e8 [ Pubmed: 35952670 DOI: doi:10.1016/j.immuni.2022.07.006 ]
- Hemagglutinin ha2 chain (26 kDa, Protein from Influenza A virus (strain A/New Zealand:South Canterbury/35/2000 H1N1))
- 310-63e6 fab, light chain (11 kDa, Protein from Homo sapiens)
- 310-63e6 fab, heavy chain (13 kDa, Protein from Homo sapiens)
- Hemagglutinin ha1 chain (35 kDa, Protein from Influenza A virus (strain A/New Zealand:South Canterbury/35/2000 H1N1))
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Structure of h1 nc99 influenza hemagglutinin bound to 310-63e6 fab (Complex from Influenza A virus)
Structure of H1 influenza hemagglutinin bound to Fab 310-39G10
Resolution: 3.37 Å
EM Method: Single-particle
Fitted PDBs: 7sco
Q-score: 0.451
Sangesland M, Torrents de la Pena A, Boyoglu-Barnum S, Ronsard L, Mohamed FAN, Moreno TB, Barnes RM, Rohrer D, Lonberg N, Ghebremichael M, Kanekiyo M, Ward A, Lingwood D
Immunity (2022) 55 pp. 1693-1709.e8 [ Pubmed: 35952670 DOI: doi:10.1016/j.immuni.2022.07.006 ]
- 310-39g10 fab, light chain (11 kDa, Protein from Homo sapiens)
- Hemagglutinin ha2 chain (26 kDa, Protein from Influenza A virus (strain A/New Zealand:South Canterbury/35/2000 H1N1))
- 310-39g10 fab, heavy chain (12 kDa, Protein from Homo sapiens)
- Structure of h1 nc99 influenza hemagglutinin bound to fab 310-39g10 (Complex from Influenza A virus)
- Hemagglutinin ha1 chain (35 kDa, Protein from Influenza A virus (strain A/New Zealand:South Canterbury/35/2000 H1N1))
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Rabies virus glycoprotein pre-fusion trimer in complex with neutralizing antibody RVA122
Resolution: 3.39 Å
EM Method: Single-particle
Fitted PDBs: 7u9g
Q-score: 0.459
Callaway HM, Zyla D, Larrous F, de Melo GD, Hastie KM, Avalos RD, Agarwal A, Corti D, Bourhy H, Saphire EO
Sci Adv (2022) 8 pp. eabp9151-eabp9151 [ DOI: doi:10.1126/sciadv.abp9151 Pubmed: 35714192 ]
- Rva122 fab light chain (23 kDa, Protein from Homo sapiens)
- Rabies virus glycoprotein pre-fusion trimer in complex with neutralizing antibody rva122 (Complex)
- Glycoprotein (49 kDa, Protein from Rabies virus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Rva122 fab heavy chain (28 kDa, Protein from Homo sapiens)
Cryo-EM structure of NTD-directed neutralizing antibody LP5 Fab in complex with SARS-CoV-2 S2P spike
Resolution: 4.46 Å
EM Method: Single-particle
Fitted PDBs: 7mxp
Q-score: 0.192
Madan B, Reddem ER, Wang P, Casner RG, Nair MS, Huang Y, Fahad AS, de Souza MO, Banach BB, Lopez Acevedo SN, Pan X, Nimrania R, Teng IT, Bahna F, Zhou T, Zhang B, Yin MT, Ho DD, Kwong PD, Shapiro L, DeKosky BJ
Aiche J (2021) 67 pp. e17440-e17440 [ Pubmed: 34898670 DOI: doi:10.1002/aic.17440 ]
- Lp5 fab light chain (11 kDa, Protein from Homo sapiens)
- Lp5 heavy chain, lp5 light chain (Complex from Homo sapiens)
- Lp5 fab heavy chain (13 kDa, Protein from Homo sapiens)
- Prefusion sars-cov-2 spike glycoprotein in complex with lp5 fab (Complex)
- Spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of HIV-1 Env trimer BG505 SOSIP.664 in complex with CD4bs antibody Ab1573
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 7tfo
Q-score: 0.359
Yang Z, Dam KA, Bridges MD, Hoffmann MAG, DeLaitsch AT, Gristick HB, Escolano A, Gautam R, Martin MA, Nussenzweig MC, Hubbell WL, Bjorkman PJ
Nat Commun (2022) 13 pp. 732-732 [ DOI: doi:10.1038/s41467-022-28424-3 Pubmed: 35136084 ]
- Bg505 sosip.664 in complex with cd4bs antibody ab1573 (Complex from Macaca mulatta)
- Envelope glycoprotein bg505 sosip.664 - gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Envelope glycoprotein bg505 sosip.664 - gp120 (54 kDa, Protein from Human immunodeficiency virus 1)
- Cd4 binding site antibody ab1573 - fab heavy chain (24 kDa, Protein from Macaca mulatta)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cd4 binding site antibody ab1573 - fab light chain (23 kDa, Protein from Macaca mulatta)