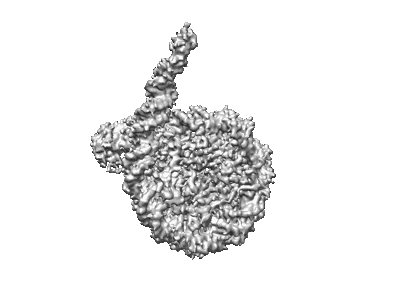
Cryo-EM structure of nucleosome containing Widom603 DNA
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 8rgm
Q-score: 0.352
Motorin NA, Afonin D, Armeev GA, Moiseenko A, Zhao L, Vasiliev V, Oleinikov P, Shaytan A, Shi X, Studitsky V, Sokolova O
To Be Published
- Histone h2a type 1-b/e (14 kDa, Protein from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Widom 603 dna sequence (44 kDa, DNA from synthetic construct)
- Widom 603 dna sequence (44 kDa, DNA from synthetic construct)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Histone h2b type 1-j (13 kDa, Protein from Homo sapiens)
- Human nucleosome containing widom 603 dna sequence (Complex from Homo sapiens)
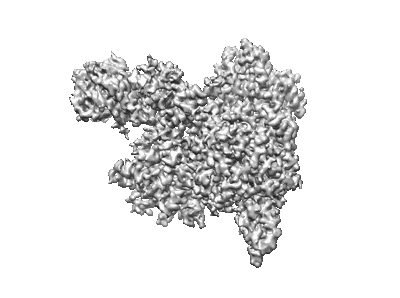
Human OGG1 bound to a nucleosome core particle with 8-oxodGuo lesion at SHL6.0
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 9eoz
Q-score: 0.465
Ren M, Gut F, Fan Y, Ma J, Shan X, Yikilmazsoy A, Likhodeeva M, Hopfner KP, Zhou C
Nat Commun (2024) 15 pp. 9407-9407 [ DOI: doi:10.1038/s41467-024-53811-3 Pubmed: 39477986 ]
- Widom 601 dna (145-mer) (44 kDa, DNA from synthetic construct)
- Histone h2a type 1-c (14 kDa, Protein from Homo sapiens)
- Widom 601 dna (145-mer) (45 kDa, DNA from synthetic construct)
- Nucleosome core particle (Complex from Homo sapiens)
- Histone h3.3 (15 kDa, Protein from Homo sapiens)
- Histone h2b type 1-c/e/f/g/i (13 kDa, Protein from Homo sapiens)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Widom 601 dna (Complex from synthetic construct)
- Binary complex of human 8-oxoguanine glycosylase with nucleosome core particle (Complex)
- N-glycosylase/dna lyase (40 kDa, Protein from Homo sapiens)
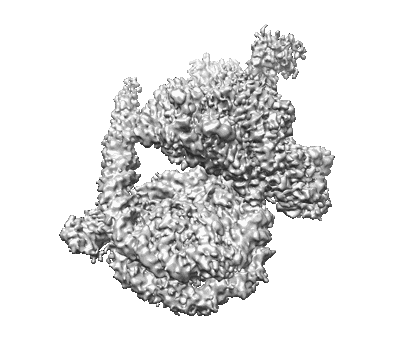
Structural basis for the nucleosome binding and chromatin compaction by the linker histone H5
Structural basis for the nucleosome binding and chromatin compaction by the linker histone H5
Structural basis for the linker histone H5-nucleosome binding and chromatin compaction
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8xjv
Q-score: 0.424
Li W, Hu J, Song F, Yu J, Peng X, Zhang S, Wang L, Hu M, Liu JC, Wei Y, Xiao X, Li Y, Li D, Wang H, Zhou BR, Dai L, Mou Z, Zhou M, Zhang H, Zhou Z, Zhang H, Bai Y, Zhou JQ, Li W, Li G, Zhu P
Cell Res (2024) 34 pp. 707-724 [ DOI: doi:10.1038/s41422-024-01009-z Pubmed: 39103524 ]
- Histone h2a (14 kDa, Protein from Xenopus laevis)
- Histone h4 (Complex from Xenopus laevis)
- Histone h2b (Complex from Xenopus laevis)
- Linker histone h5 (Complex from Gallus gallus)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Histone h5 (21 kDa, Protein from Gallus gallus)
- Dna (660 kDa, DNA from synthetic construct)
- 12x177bp-h5 chromatin (Complex)
- Dna (651 kDa, DNA from synthetic construct)
- Histone h3.1 (Complex from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Widom 601 dna (Complex from synthetic construct)
- Histone h2a (Complex from Xenopus laevis)
Eaf3 CHD domain bound to the nucleosome
Resolution: 3.58 Å
EM Method: Single-particle
Fitted PDBs: 8ihm
Q-score: 0.434
Zhang Y, Xu M, Wang P, Zhou J, Wang G, Han S, Cai G, Wang X
Cell Res (2023) 33 pp. 971-974 [ DOI: doi:10.1038/s41422-023-00884-2 Pubmed: 37845487 ]
- Rco1-abr (2 kDa, Protein from Saccharomyces cerevisiae)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Dna (165-mer) (51 kDa, DNA from Xenopus laevis)
- Dna (164-mer) (50 kDa, DNA from Xenopus laevis)
- Chromatin modification-related protein eaf3 (13 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Eaf3 chd domain bound to the nucleosome (Complex from Xenopus laevis)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
Cryo-EM structure of the Rpd3S core complex
Resolution: 3.37 Å
EM Method: Single-particle
Fitted PDBs: 8ihn
Q-score: 0.48
Zhang Y, Xu M, Wang P, Zhou J, Wang G, Han S, Cai G, Wang X
Cell Res (2023) 33 pp. 971-974 [ DOI: doi:10.1038/s41422-023-00884-2 Pubmed: 37845487 ]
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Eaf3 chd domain bound to the nucleosome (Complex from Xenopus laevis)
- Calcium ion (40 Da, Ligand)
- Rco1 isoform 1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (2 kDa, Protein from Xenopus laevis)
Resolution: 3.72 Å
EM Method: Single-particle
Fitted PDBs: 8iht
Q-score: 0.345
Zhang Y, Xu M, Wang P, Zhou J, Wang G, Han S, Cai G, Wang X
Cell Res (2023) 33 pp. 971-974 [ DOI: doi:10.1038/s41422-023-00884-2 Pubmed: 37845487 ]
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Dna (165-mer) (51 kDa, DNA from Xenopus laevis)
- Dna (164-mer) (50 kDa, DNA from Xenopus laevis)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Eaf3 chd domain bound to the nucleosome (Complex from Xenopus laevis)
- Calcium ion (40 Da, Ligand)
- Rco1 isoform 1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
Cryo-EM structure of SSX1 bound to the H2AK119Ub nucleosome at a resolution of 3.05 angstrom
Resolution: 3.05 Å
EM Method: Single-particle
Fitted PDBs: 8hqy
Q-score: 0.47
Tong Z, Ai H, Xu Z, He K, Chu GC, Shi Q, Deng Z, Xue Q, Sun M, Du Y, Liang L, Li JB, Pan M, Liu L
Nat Struct Mol Biol (2024) 31 pp. 300-310 [ DOI: doi:10.1038/s41594-023-01141-1 Pubmed: 38177667 ]
- Ssx1-h2ak119ub nucleosome complex (290 kDa, Complex from Homo sapiens)
- Histone h3 (11 kDa, Protein from Homo sapiens)
- Polyubiquitin-b (fragment) (8 kDa, Protein from Homo sapiens)
- Dna (137-mer) (41 kDa, DNA from Homo sapiens)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- Histone h4 (9 kDa, Protein from Homo sapiens)
- Dna (136-mer) (42 kDa, DNA from Homo sapiens)
- Histone h2a type 1-b/e (11 kDa, Protein from Homo sapiens)
- Protein ssx2 (2 kDa, Protein from Homo sapiens)
- Histone h2a type 1-b/e (11 kDa, Protein from Homo sapiens)
- Histone h4 (fragment) (9 kDa, Protein from Homo sapiens)
Cryo-EM structure of SSX1 bound to the unmodified nucleosome at a resolution of 3.02 angstrom
Resolution: 3.02 Å
EM Method: Single-particle
Fitted PDBs: 8hr1
Q-score: 0.532
Tong Z, Ai H, Xu Z, He K, Chu GC, Shi Q, Deng Z, Xue Q, Sun M, Du Y, Liang L, Li JB, Pan M, Liu L
Nat Struct Mol Biol (2024) 31 pp. 300-310 [ DOI: doi:10.1038/s41594-023-01141-1 Pubmed: 38177667 ]
- Histone h3 (11 kDa, Protein from Homo sapiens)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- Ssx1 (1 kDa, Protein from Homo sapiens)
- Dna (147-mer) (45 kDa, DNA from Homo sapiens)
- Histone h4 (9 kDa, Protein from Homo sapiens)
- Histone h2a type 1-b/e (11 kDa, Protein from Homo sapiens)
- Ssx1-unmodified nucleosome complex (280 kDa, Complex from Homo sapiens)
- Dna (147-mer) (45 kDa, DNA from Homo sapiens)