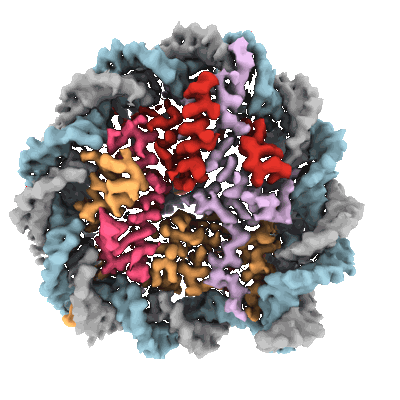
Cryo-EM structure of native SWR1 bound to nucleosome (composite structure)
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 9b1e
Q-score: 0.191
Louder RK, Park G, Ye Z, Cha JS, Gardner AM, Lei Q, Ranjan A, Hollmuller E, Stengel F, Pugh BF, Wu C
Cell (2024) 187 pp. 6849-6864.e18 [ Pubmed: 39357520 DOI: doi:10.1016/j.cell.2024.09.007 ]
- Dna (214-mer) (66 kDa, DNA from synthetic construct)
- Vacuolar protein sorting-associated protein 72 (90 kDa, Protein from Saccharomyces cerevisiae W303)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Ruvb-like protein 1 (91 kDa, Protein from Saccharomyces cerevisiae W303)
- Histone h2a (14 kDa, Protein from Saccharomyces cerevisiae)
- Dna (214-mer) (65 kDa, DNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Native swr1 bound to nucleosome. (1 MDa, Complex from Saccharomyces cerevisiae)
- Helicase swr1 (178 kDa, Protein from Saccharomyces cerevisiae W303)
- Histone h4 (11 kDa, Protein from Drosophila melanogaster)
- Zinc ion (65 Da, Ligand)
- Actin-like protein arp6 (50 kDa, Protein from Saccharomyces cerevisiae W303)
- Beryllium trifluoride ion (66 Da, Ligand)
- Swr1-complex protein 3 (72 kDa, Protein from Saccharomyces cerevisiae W303)
- Vacuolar protein sorting-associated protein 71 (32 kDa, Protein from Saccharomyces cerevisiae W303)
- Ruvb-like protein 2 (51 kDa, Protein from Saccharomyces cerevisiae W303)
- Histone h3 (15 kDa, Protein from Drosophila melanogaster)
- Native swr1 complex (Complex from Saccharomyces cerevisiae)
- Histone h2b (14 kDa, Protein from Saccharomyces cerevisiae)
Structural basis for the linker histone H5-nucleosome binding and chromatin compaction
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8xjv
Q-score: 0.424
Li W, Hu J, Song F, Yu J, Peng X, Zhang S, Wang L, Hu M, Liu JC, Wei Y, Xiao X, Li Y, Li D, Wang H, Zhou BR, Dai L, Mou Z, Zhou M, Zhang H, Zhou Z, Zhang H, Bai Y, Zhou JQ, Li W, Li G, Zhu P
Cell Res (2024) 34 pp. 707-724 [ DOI: doi:10.1038/s41422-024-01009-z Pubmed: 39103524 ]
- Histone h2a (14 kDa, Protein from Xenopus laevis)
- Histone h4 (Complex from Xenopus laevis)
- Histone h2b (Complex from Xenopus laevis)
- Linker histone h5 (Complex from Gallus gallus)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Histone h5 (21 kDa, Protein from Gallus gallus)
- Dna (660 kDa, DNA from synthetic construct)
- 12x177bp-h5 chromatin (Complex)
- Dna (651 kDa, DNA from synthetic construct)
- Histone h3.1 (Complex from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Widom 601 dna (Complex from synthetic construct)
- Histone h2a (Complex from Xenopus laevis)
P400 subcomplex of the native human TIP60 complex
Yang Z, Mameri A, Cattoglio C, Lachance C, Florez Ariza AJ, Luo J, Humbert J, Sudarshan D, Banerjea A, Galloy M, Fradet-Turcotte A, Lambert JP, Ranish JA, Cote J, Nogales E
Science (2024) 385 pp. eadl5816-eadl5816 [ Pubmed: 39088653 DOI: doi:10.1126/science.adl5816 ]
- Dna methyltransferase 1-associated protein 1 (53 kDa, Protein from Homo sapiens)
- Histone h2a.z (13 kDa, Protein from Homo sapiens)
- Reconstituted p400 subcomplex of the human tip60 complex (Complex from Homo sapiens)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- E1a-binding protein p400 (343 kDa, Protein from Homo sapiens)
- Ruvb-like 2 (51 kDa, Protein from Homo sapiens)
- Actin, cytoplasmic 1 (41 kDa, Protein from Homo sapiens)
- Vacuolar protein sorting-associated protein 72 homolog (40 kDa, Protein from Homo sapiens)
- Histone h2b type 1-b (13 kDa, Protein from Homo sapiens)
- Ruvb-like 1 (50 kDa, Protein from Homo sapiens)
- Actin-like protein 6a (47 kDa, Protein from Homo sapiens)
- Enhancer of polycomb homolog 1 (93 kDa, Protein from Homo sapiens)
Resolution: 3.27 Å
EM Method: Single-particle
Fitted PDBs: 8x7i
Q-score: 0.443
Ai H, Tong Z, Deng Z, Shi Q, Tao S, Liang J, Sun M, Wu X, Zheng Q, Liang L, Li JB, Gao S, Tian C, Liu L, Pan M
bioRxiv (2024) [ DOI: doi:10.1101/2024.01.02.573964 ]
- Histone h2a type 1-b/e (11 kDa, Protein from Homo sapiens)
- Rnf168/ubch5c-ub/nucleosomes determined by intein-based e2-ub-ncp conjugation strategy (Complex from Homo sapiens)
- Histone h2a type 1-b/e (12 kDa, Protein from Homo sapiens)
- E3 ubiquitin-protein ligase rnf168 (12 kDa, Protein from Homo sapiens)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- Ubiquitin-conjugating enzyme e2 d3 (16 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Dna (147-mer) (45 kDa, DNA from Homo sapiens)
- Histone h4 (9 kDa, Protein from Homo sapiens)
- Histone h3.2 (11 kDa, Protein from Homo sapiens)
- Ubiquitin (8 kDa, Protein from Homo sapiens)
- Dna (147-mer) (45 kDa, DNA from Homo sapiens)
Resolution: 3.39 Å
EM Method: Single-particle
Fitted PDBs: 8x7j
Q-score: 0.391
Ai H, Tong Z, Deng Z, Shi Q, Tao S, Liang J, Sun M, Wu X, Zheng Q, Liang L, Li JB, Gao S, Tian C, Liu L, Pan M
bioRxiv (2024) [ DOI: doi:10.1101/2024.01.02.573964 ]
- E3 ubiquitin-protein ligase rnf168 (12 kDa, Protein from Homo sapiens)
- Histone h2a type 1-b/e (11 kDa, Protein from Homo sapiens)
- Histone h3.2 (11 kDa, Protein from Homo sapiens)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- Histone h2a type 1-b/e (11 kDa, Protein from Homo sapiens)
- Ubiquitin-conjugating enzyme e2 d3 (16 kDa, Protein from Homo sapiens)
- Dna (144-mer) (44 kDa, DNA from Homo sapiens)
- Dna (144-mer) (44 kDa, DNA from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Ubiquitin (8 kDa, Protein from Homo sapiens)
- Rnf168/ubch5c-ub/nucleosomes complex determined by activity-based chemical trapping strategy (Complex from Homo sapiens)
- Histone h4 (9 kDa, Protein from Homo sapiens)
Resolution: 3.27 Å
EM Method: Single-particle
Fitted PDBs: 8x7k
Q-score: 0.423
Ai H, Tong Z, Deng Z, Shi Q, Tao S, Liang J, Sun M, Wu X, Zheng Q, Liang L, Li JB, Gao S, Tian C, Liu L, Pan M
bioRxiv (2024) [ DOI: doi:10.1101/2024.01.02.573964 ]
- E3 ubiquitin-protein ligase rnf168 (12 kDa, Protein from Homo sapiens)
- Histone h3.2 (11 kDa, Protein from Homo sapiens)
- Histone h2a type 1-b/e (11 kDa, Protein from Homo sapiens)
- Dna (143-mer) (44 kDa, DNA from Homo sapiens)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- Cryo-em structures of rnf168/ubch5c-ub in complex with h2ak13ub nucleosomes determined by activity-based chemical trapping strategy (adjacent h2ak13/15 dual-monoubiquitination) (Complex from Homo sapiens)
- Histone h2a type 1-b/e (11 kDa, Protein from Homo sapiens)
- Ubiquitin-conjugating enzyme e2 d3 (16 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Dna (143-mer) (43 kDa, DNA from Homo sapiens)
- Histone h4 (9 kDa, Protein from Homo sapiens)
CryoEM structure of DIM2-HP1-H3K9me3-DNA complex
Resolution: 2.79 Å
EM Method: Single-particle
Fitted PDBs: 9baq
Q-score: 0.542
Shao Z, Lu J, Khudaverdyan N, Song J
Nat Commun (2024) 15 pp. 6815-6815 [ DOI: doi:10.1038/s41467-024-51246-4 Pubmed: 39122718 ]
- Dna (5'-d(*ap*gp*tp*ap*gp*gp*ap*gp*gp*ap*gp*gp*ap*gp*tp*ap*gp*t)-3') (5 kDa, DNA from Neurospora crassa)
- Zinc ion (65 Da, Ligand)
- Dna (cytosine-5-)-methyltransferase (139 kDa, Protein from Neurospora crassa)
- Dna (5'-d(*ap*cp*tp*ap*cp*t)-r(p*(pyo))-d(p*cp*tp*cp*cp*tp*cp*cp*tp*ap*cp*t)-3') (5 kDa, DNA from Neurospora crassa)
- Histone h3.2 (2 kDa, Protein from Neurospora crassa)
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- Heterochromatin protein one (30 kDa, Protein from Neurospora crassa)
- Dim2-hp1-h3k9m3-dna (Complex from Neurospora crassa)
Complex of DDM1-nucleosome(H2A) complex with DDM1 bound to SHL2
Resolution: 3.17 Å
EM Method: Single-particle
Fitted PDBs: 8kcb
Q-score: 0.38
Zhang H, Gu Z, Zeng Y, Zhang Y
Structure (2024) 32 pp. 1222-1230.e4 [ DOI: doi:10.1016/j.str.2024.05.013 Pubmed: 38870940 ]
- Dna (170-mer) (52 kDa, DNA from synthetic construct)
- Beryllium trifluoride ion (66 Da, Ligand)
- Histone h2b.10 (15 kDa, Protein from Arabidopsis thaliana)
- Complex of ddm1-nucleosome(h2a.w) complex with ddm1 bound to shl2 (Complex from Arabidopsis thaliana)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Histone h4 (11 kDa, Protein from Arabidopsis thaliana)
- Histone h3.1 (15 kDa, Protein from Arabidopsis thaliana)
- Histone h2a.6 (13 kDa, Protein from Arabidopsis thaliana)
- Dna (170-mer) (52 kDa, DNA from synthetic construct)
- Atp-dependent dna helicase ddm1 (86 kDa, Protein from Arabidopsis thaliana)
Complex of DDM1-nucleosome(H2A.W) complex with DDM1 bound to SHL2
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8kcc
Q-score: 0.387
Zhang H, Gu Z, Zeng Y, Zhang Y
Structure (2024) 32 pp. 1222-1230.e4 [ DOI: doi:10.1016/j.str.2024.05.013 Pubmed: 38870940 ]
- Dna (170-mer) (52 kDa, DNA from synthetic construct)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Histone h2b.10 (15 kDa, Protein from Arabidopsis thaliana)
- Complex of ddm1-nucleosome(h2a.w) complex with ddm1 bound to shl2 (Complex from Arabidopsis thaliana)
- Beryllium trifluoride ion (66 Da, Ligand)
- Histone h4 (11 kDa, Protein from Arabidopsis thaliana)
- Histone h3.1 (15 kDa, Protein from Arabidopsis thaliana)
- Dna (170-mer) (52 kDa, DNA from synthetic construct)
- Atp-dependent dna helicase ddm1 (86 kDa, Protein from Arabidopsis thaliana)
- Probable histone h2a.7 (15 kDa, Protein from Arabidopsis thaliana)
Human canonical 601 DNA nucleosome
Resolution: 3.35 Å
EM Method: Single-particle
Fitted PDBs: 8jbx
Q-score: 0.43
Ding D, Pang MYH, Deng M, Nguyen TT, Liu Y, Sun X, Xu Z, Zhang Y, Zhai Y, Yan Y, Ishibashi T
Nucleic Acids Res (2024) 52 pp. 11612-11625 [ DOI: doi:10.1093/nar/gkae825 Pubmed: 39329259 ]
- Dna (147-mer) (45 kDa, DNA from synthetic construct)
- Human canonical histone and 601 dna (Complex)
- 601 dna (Complex)
- Dna (147-mer) (45 kDa, DNA from synthetic construct)
- Human canonical histone (Complex from Homo sapiens)
- Histone h2a type 1-b/e (14 kDa, Protein from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Histone h2b type 1-c/e/f/g/i (13 kDa, Protein from Homo sapiens)
- Histone h4 (11 kDa, Protein from Homo sapiens)