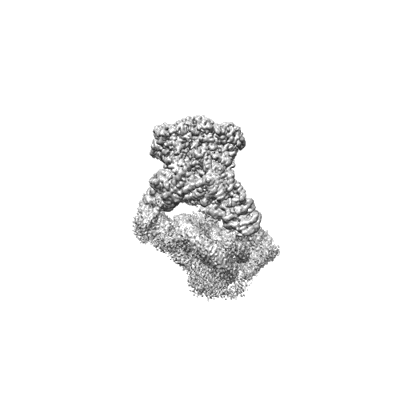
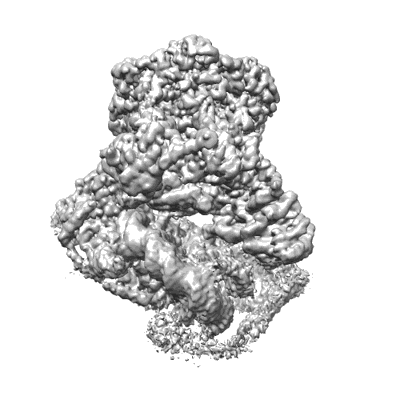
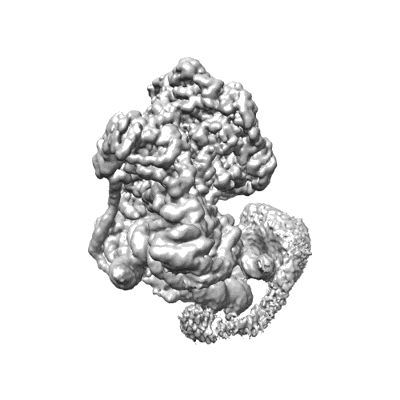
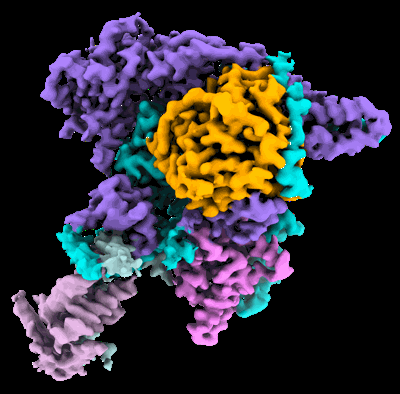
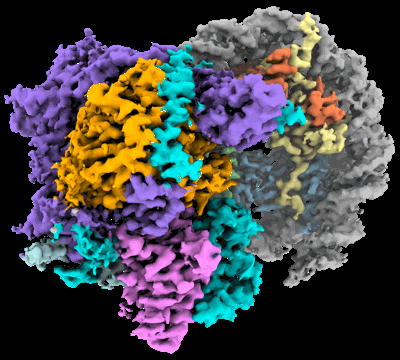
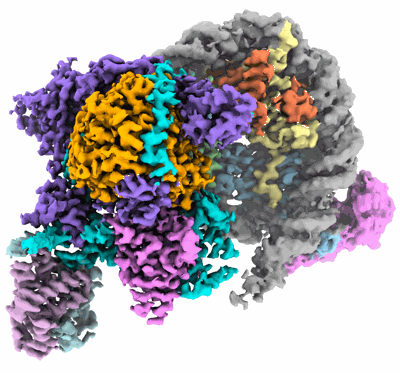
Structural basis for the linker histone H5-nucleosome binding and chromatin compaction
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8xjv
Q-score: 0.424
Li W, Hu J, Song F, Yu J, Peng X, Zhang S, Wang L, Hu M, Liu JC, Wei Y, Xiao X, Li Y, Li D, Wang H, Zhou BR, Dai L, Mou Z, Zhou M, Zhang H, Zhou Z, Zhang H, Bai Y, Zhou JQ, Li W, Li G, Zhu P
Cell Res (2024) 34 pp. 707-724 [ DOI: doi:10.1038/s41422-024-01009-z Pubmed: 39103524 ]
- Histone h2a (14 kDa, Protein from Xenopus laevis)
- Histone h4 (Complex from Xenopus laevis)
- Histone h2b (Complex from Xenopus laevis)
- Linker histone h5 (Complex from Gallus gallus)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Histone h5 (21 kDa, Protein from Gallus gallus)
- Dna (660 kDa, DNA from synthetic construct)
- 12x177bp-h5 chromatin (Complex)
- Dna (651 kDa, DNA from synthetic construct)
- Histone h3.1 (Complex from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Widom 601 dna (Complex from synthetic construct)
- Histone h2a (Complex from Xenopus laevis)
Structure of nucleosome-bound SRCAP-C in the apo state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8x15
Q-score: 0.218
Yu J, Sui F, Gu F, Li W, Yu Z, Wang Q, He S, Wang L, Xu Y
Cell Discov (2024) 10 pp. 15-15 [ Pubmed: 38331872 DOI: doi:10.1038/s41421-023-00640-1 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Ruvb-like 2 (51 kDa, Protein from Homo sapiens)
- Dna (137-mer) (42 kDa, DNA from synthetic construct)
- Structure of nucleosome-bound srcap-c in the apo state (Complex from Homo sapiens)
- Actin-like protein 6a (47 kDa, Protein from Homo sapiens)
- Vacuolar protein sorting-associated protein 72 homolog (40 kDa, Protein from Homo sapiens)
- Ruvb-like 1 (50 kDa, Protein from Homo sapiens)
- Dna methyltransferase 1-associated protein 1 (53 kDa, Protein from Homo sapiens)
- Dna (137-mer) (41 kDa, DNA from synthetic construct)
- Actin-related protein 6 (45 kDa, Protein from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Zinc finger hit domain-containing protein 1 (17 kDa, Protein from Homo sapiens)
- Histone h2a type 1-c (14 kDa, Protein from Homo sapiens)
- Histone h2b type 1-c/e/f/g/i (13 kDa, Protein from Homo sapiens)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Helicase srcap (343 kDa, Protein from Homo sapiens)
- Yeats domain-containing protein 4 (26 kDa, Protein from Homo sapiens)
- Actin, cytoplasmic 1, n-terminally processed (41 kDa, Protein from Homo sapiens)
Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8x19
Q-score: 0.292
Yu J, Sui F, Gu F, Li W, Yu Z, Wang Q, He S, Wang L, Xu Y
Cell Discov (2024) 10 pp. 15-15 [ Pubmed: 38331872 DOI: doi:10.1038/s41421-023-00640-1 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Dna (147-mer) (45 kDa, DNA from Homo sapiens)
- Ruvb-like 2 (51 kDa, Protein from Homo sapiens)
- Actin-like protein 6a (47 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Vacuolar protein sorting-associated protein 72 homolog (40 kDa, Protein from Homo sapiens)
- Ruvb-like 1 (50 kDa, Protein from Homo sapiens)
- Dna methyltransferase 1-associated protein 1 (53 kDa, Protein from Homo sapiens)
- Actin-related protein 6 (45 kDa, Protein from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Zinc finger hit domain-containing protein 1 (17 kDa, Protein from Homo sapiens)
- Histone h2a type 1-c (14 kDa, Protein from Homo sapiens)
- Histone h2b type 1-c/e/f/g/i (13 kDa, Protein from Homo sapiens)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Beryllium trifluoride ion (66 Da, Ligand)
- Helicase srcap (343 kDa, Protein from Homo sapiens)
- Dna (147-mer) (45 kDa, DNA from Homo sapiens)
- Actin, cytoplasmic 1 (41 kDa, Protein from Homo sapiens)
- Yeats domain-containing protein 4 (26 kDa, Protein from Homo sapiens)
- Structure of nucleosome-bound srcap-c in the adp-befx-bound state (Complex from Homo sapiens)
Structure of nucleosome-bound SRCAP-C in the ADP-bound state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8x1c
Q-score: 0.297
Yu J, Sui F, Gu F, Li W, Yu Z, Wang Q, He S, Wang L, Xu Y
Cell Discov (2024) 10 pp. 15-15 [ Pubmed: 38331872 DOI: doi:10.1038/s41421-023-00640-1 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Ruvb-like 2 (51 kDa, Protein from Homo sapiens)
- Dna (147-mer) (45 kDa, DNA from synthetic construct)
- Actin-like protein 6a (47 kDa, Protein from Homo sapiens)
- Structure of nucleosome-bound srcap-c in the adp-bound state (Complex from Homo sapiens)
- Vacuolar protein sorting-associated protein 72 homolog (40 kDa, Protein from Homo sapiens)
- Ruvb-like 1 (50 kDa, Protein from Homo sapiens)
- Dna (147-mer) (45 kDa, DNA from synthetic construct)
- Dna methyltransferase 1-associated protein 1 (53 kDa, Protein from Homo sapiens)
- Actin-related protein 6 (45 kDa, Protein from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Zinc finger hit domain-containing protein 1 (17 kDa, Protein from Homo sapiens)
- Histone h2a type 1-c (14 kDa, Protein from Homo sapiens)
- Histone h2b type 1-c/e/f/g/i (13 kDa, Protein from Homo sapiens)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Helicase srcap (343 kDa, Protein from Homo sapiens)
- Actin, cytoplasmic 1 (41 kDa, Protein from Homo sapiens)
- Yeats domain-containing protein 4 (26 kDa, Protein from Homo sapiens)
Native SUV420H1 bound to 167-bp nucleosome
Resolution: 3.58 Å
EM Method: Single-particle
Fitted PDBs: 8jhg
Q-score: 0.456
Lin F, Zhang R, Shao W, Lei C, Ma M, Zhang Y, Wen Z, Li W
Cell Discov (2023) 9 pp. 120-120 [ DOI: doi:10.1038/s41421-023-00620-5 Pubmed: 38052811 ]
- Histone h4 (9 kDa, Protein from Homo sapiens)
- Histone h2a type 1-b/e (12 kDa, Protein from Homo sapiens)
- Dna (160-mer) (49 kDa, DNA from Homo sapiens)
- Dna (160-mer) (49 kDa, DNA from Homo sapiens)
- Histone-lysine n-methyltransferase kmt5b (41 kDa, Protein from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- S-adenosylmethionine (398 Da, Ligand)
- Cryo-em structure of the native suv420h1 bound to 167-bp nucleosome (246 kDa, Complex from Homo sapiens)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
Native SUV420H1 bound to 167-bp nucleosome
Resolution: 3.68 Å
EM Method: Single-particle
Fitted PDBs: 8jhf
Q-score: 0.433
Lin F, Zhang R, Shao W, Lei C, Ma M, Zhang Y, Wen Z, Li W
Cell Discov (2023) 9 pp. 120-120 [ DOI: doi:10.1038/s41421-023-00620-5 Pubmed: 38052811 ]
- Histone h4 (9 kDa, Protein from Homo sapiens)
- Dna (160-mer) (49 kDa, DNA from Homo sapiens)
- Dna (160-mer) (49 kDa, DNA from Homo sapiens)
- Histone-lysine n-methyltransferase kmt5b (41 kDa, Protein from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- S-adenosylmethionine (398 Da, Ligand)
- Cryo-em structure of the native suv420h1 bound to 167-bp nucleosome (246 kDa, Complex from Homo sapiens)
- Histone h2a.z (12 kDa, Protein from Homo sapiens)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
Cryo-EM structure of the histone deacetylase complex Rpd3S
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8hxx
Q-score: 0.453
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ Pubmed: 37798513 DOI: doi:10.1038/s41594-023-01121-5 ]
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3s histone deacetylase in complex with histone h3 tail (Complex from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Rco1 isoform 1 (78 kDa, Protein from Saccharomyces cerevisiae)
Cryo-EM structure of the histone deacetylase complex Rpd3S in complex with nucleosome
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8hxy
Q-score: 0.44
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ Pubmed: 37798513 DOI: doi:10.1038/s41594-023-01121-5 ]
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3s histone deacetylase in complex with nucleosome (680 kDa, Complex from Saccharomyces cerevisiae)
- Transcriptional regulatory protein rco1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Dna (352-mer) (108 kDa, DNA from synthetic construct)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Dna (352-mer) (109 kDa, DNA from synthetic construct)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
Cryo-EM structure of Eaf3 CHD in complex with nucleosome
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8hxz
Q-score: 0.326
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ Pubmed: 37798513 DOI: doi:10.1038/s41594-023-01121-5 ]
- Eaf3 chd in complex with nucleosome (150 kDa, Complex from Saccharomyces)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Dna (352-mer) (108 kDa, DNA from synthetic construct)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Dna (352-mer) (109 kDa, DNA from synthetic construct)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
Composite cryo-EM structure of the histone deacetylase complex Rpd3S in complex with nucleosome
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8hy0
Q-score: 0.463
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ Pubmed: 37798513 DOI: doi:10.1038/s41594-023-01121-5 ]
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Dna (352-mer) (108 kDa, DNA from synthetic construct)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3s histone deacetylase in complex with nucleosome (800 kDa, Complex from Saccharomyces cerevisiae)
- Dna (352-mer) (109 kDa, DNA from synthetic construct)
- Rco1 isoform 1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Histone h2b (13 kDa, Protein from Xenopus laevis)