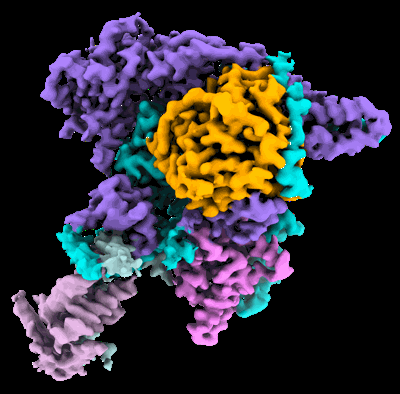
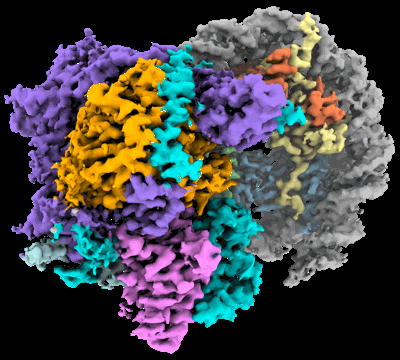
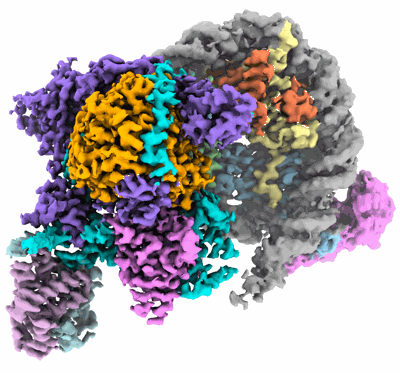
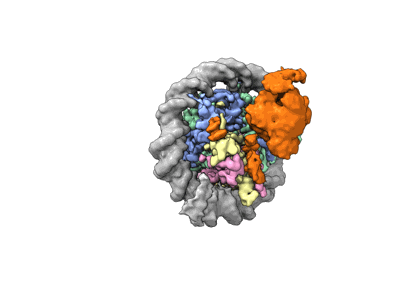
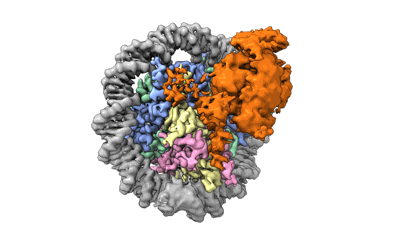
Cryo-EM structure of DNMT3A1 UDR in complex with H2AK119Ub-nucleosome
Gretarsson KH, Abini-Agbomson S, Gloor SL, Weinberg DN, McCuiston JL, Kumary VUS, Hickman AR, Sahu V, Lee R, Xu X, Lipieta N, Flashner S, Adeleke OA, Popova IK, Taylor HF, Noll K, Windham CL, Maryanski DN, Venters BJ, Nakagawa H, Keogh MC, Armache KJ, Lu C
Sci Adv (2024) 10 pp. eadp0975-eadp0975 [ DOI: doi:10.1126/sciadv.adp0975 Pubmed: 39196936 ]
- Histone h2a (14 kDa, Protein from Xenopus laevis)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Dna (146-mer) (44 kDa, DNA from Escherichia coli 'BL21-Gold(DE3)pLysS AG')
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Dnmt3a1 udr in complex with h2ak119ub-nucleosome (Complex from Xenopus laevis)
- Dna (146-mer) (45 kDa, DNA from Escherichia coli 'BL21-Gold(DE3)pLysS AG')
- Dna (cytosine-5)-methyltransferase 3a (9 kDa, Protein from Homo sapiens)
CryoEM structure of DIM2-HP1-H3K9me3-DNA complex
Resolution: 2.79 Å
EM Method: Single-particle
Fitted PDBs: 9baq
Q-score: 0.542
Shao Z, Lu J, Khudaverdyan N, Song J
Nat Commun (2024) 15 pp. 6815-6815 [ DOI: doi:10.1038/s41467-024-51246-4 Pubmed: 39122718 ]
- Dna (5'-d(*ap*gp*tp*ap*gp*gp*ap*gp*gp*ap*gp*gp*ap*gp*tp*ap*gp*t)-3') (5 kDa, DNA from Neurospora crassa)
- Zinc ion (65 Da, Ligand)
- Dna (cytosine-5-)-methyltransferase (139 kDa, Protein from Neurospora crassa)
- Dna (5'-d(*ap*cp*tp*ap*cp*t)-r(p*(pyo))-d(p*cp*tp*cp*cp*tp*cp*cp*tp*ap*cp*t)-3') (5 kDa, DNA from Neurospora crassa)
- Histone h3.2 (2 kDa, Protein from Neurospora crassa)
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- Heterochromatin protein one (30 kDa, Protein from Neurospora crassa)
- Dim2-hp1-h3k9m3-dna (Complex from Neurospora crassa)
Cryo-EM structure of human DNMT3A UDR bound to H2AK119ub1-modified nucleosome
Resolution: 3.23 Å
EM Method: Single-particle
Fitted PDBs: 8u5h
Q-score: 0.443
Chen X, Guo Y, Zhao T, Lu J, Fang J, Wang Y, Wang GG, Song J
Nat Commun (2024) 15 pp. 6217-6217 [ Pubmed: 39043678 DOI: doi:10.1038/s41467-024-50526-3 ]
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Dnmt3a n-terminal domain and h2ak119ub nucleosome complex (Complex from Homo sapiens)
- Nucleosome dna (55 kDa, DNA from Homo sapiens)
- Nucleosome dna (55 kDa, DNA from Homo sapiens)
- Ubiquitin (8 kDa, Protein from Homo sapiens)
- Histone h3.3c (15 kDa, Protein from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Histone h2a (14 kDa, Protein from Xenopus laevis)
- Dna (cytosine-5)-methyltransferase 3a (11 kDa, Protein from Homo sapiens)
Cryo-EM structure of the histone deacetylase complex Rpd3S
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8hxx
Q-score: 0.453
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ Pubmed: 37798513 DOI: doi:10.1038/s41594-023-01121-5 ]
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3s histone deacetylase in complex with histone h3 tail (Complex from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Rco1 isoform 1 (78 kDa, Protein from Saccharomyces cerevisiae)
Cryo-EM structure of the histone deacetylase complex Rpd3S in complex with nucleosome
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8hxy
Q-score: 0.44
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ Pubmed: 37798513 DOI: doi:10.1038/s41594-023-01121-5 ]
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3s histone deacetylase in complex with nucleosome (680 kDa, Complex from Saccharomyces cerevisiae)
- Transcriptional regulatory protein rco1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Dna (352-mer) (108 kDa, DNA from synthetic construct)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Dna (352-mer) (109 kDa, DNA from synthetic construct)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
Cryo-EM structure of Eaf3 CHD in complex with nucleosome
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8hxz
Q-score: 0.326
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ Pubmed: 37798513 DOI: doi:10.1038/s41594-023-01121-5 ]
- Eaf3 chd in complex with nucleosome (150 kDa, Complex from Saccharomyces)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Dna (352-mer) (108 kDa, DNA from synthetic construct)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Dna (352-mer) (109 kDa, DNA from synthetic construct)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
Composite cryo-EM structure of the histone deacetylase complex Rpd3S in complex with nucleosome
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8hy0
Q-score: 0.463
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ Pubmed: 37798513 DOI: doi:10.1038/s41594-023-01121-5 ]
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Dna (352-mer) (108 kDa, DNA from synthetic construct)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3s histone deacetylase in complex with nucleosome (800 kDa, Complex from Saccharomyces cerevisiae)
- Dna (352-mer) (109 kDa, DNA from synthetic construct)
- Rco1 isoform 1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
Cryo-EM structure of the histone deacetylase complex Rpd3S in complex with di-nucleosome
Resolution: 7.6 Å
EM Method: Single-particle
Fitted PDBs: 8jho
Q-score: 0.067
Nat Struct Mol Biol (2023) 30 pp. 1893-1901 [ Pubmed: 37798513 DOI: doi:10.1038/s41594-023-01121-5 ]
- Di-nucleosome template foward (107 kDa, DNA from synthetic construct)
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Di-nucleosome (400 MDa, Complex from Xenopus laevis)
- Zinc ion (65 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Di-nucleosome template reverse (108 kDa, DNA from synthetic construct)
- The rpd3s hdac complex (Complex from Saccharomyces cerevisiae)
- Rco1 isoform 1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3s histone deacetylase in complex with di-nucleosome (550 MDa, Complex)
- Histone h2b (13 kDa, Protein from Xenopus laevis)
Catalytic and non-catalytic mechanisms of histone H4 lysine 20 methyltransferase SUV420H1
Resolution: 3.37 Å
EM Method: Single-particle
Fitted PDBs: 8t9h
Q-score: 0.393
Abini-Agbomson S, Gretarsson K, Shih RM, Hsieh L, Lou T, De Ioannes P, Vasilyev N, Lee R, Wang M, Simon MD, Armache JP, Nudler E, Narlikar G, Liu S, Lu C, Armache KJ
Mol Cell (2023) 83 pp. 2872-2883.e7 [ DOI: doi:10.1016/j.molcel.2023.07.020 Pubmed: 37595555 ]
- Histone h2a (14 kDa, Protein from Xenopus laevis)
- Dna (146-mer) (44 kDa, DNA from Escherichia coli 'BL21-Gold(DE3)pLysS AG')
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Suv420h1-h2a.z nucleosome complex (Complex from Xenopus laevis)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Dna (146-mer) (45 kDa, DNA from Escherichia coli 'BL21-Gold(DE3)pLysS AG')
- Histone-lysine n-methyltransferase kmt5b (44 kDa, Protein from Homo sapiens)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
Catalytic and non-catalytic mechanisms of histone H4 lysine 20 methyltransferase SUV420H1
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 8t9f
Q-score: 0.516
Abini-Agbomson S, Gretarsson K, Shih RM, Hsieh L, Lou T, De Ioannes P, Vasilyev N, Lee R, Wang M, Simon MD, Armache JP, Nudler E, Narlikar G, Liu S, Lu C, Armache KJ
Mol Cell (2023) 83 pp. 2872-2883.e7 [ DOI: doi:10.1016/j.molcel.2023.07.020 Pubmed: 37595555 ]
- Dna (122-mer) (44 kDa, DNA from Escherichia coli 'BL21-Gold(DE3)pLysS AG')
- Histone h2a.z (13 kDa, Protein from Homo sapiens)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Suv420h1-h2a.z nucleosome complex (Complex from Xenopus laevis)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- S-adenosylmethionine (398 Da, Ligand)
- Histone-lysine n-methyltransferase kmt5b (44 kDa, Protein from Homo sapiens)
- Dna (122-mer) (45 kDa, DNA from Escherichia coli 'BL21-Gold(DE3)pLysS AG')
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)