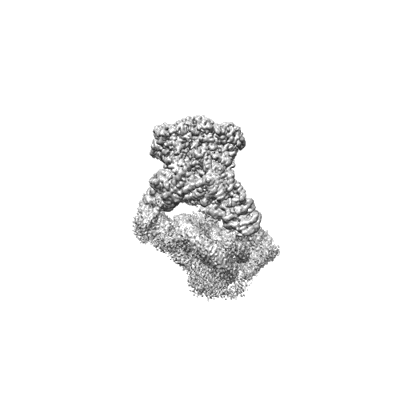
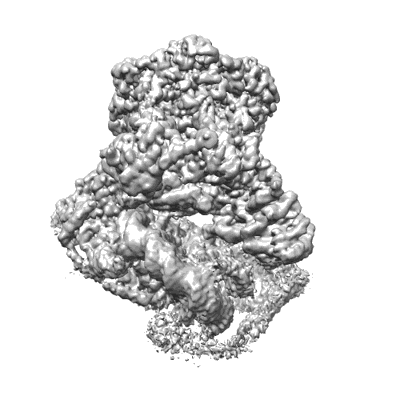
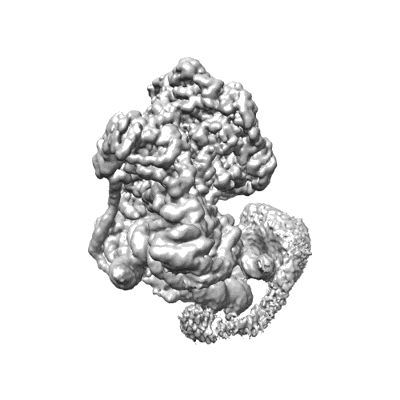
Structure of nucleosome-bound SRCAP-C in the apo state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8x15
Q-score: 0.218
Yu J, Sui F, Gu F, Li W, Yu Z, Wang Q, He S, Wang L, Xu Y
Cell Discov (2024) 10 pp. 15-15 [ DOI: doi:10.1038/s41421-023-00640-1 Pubmed: 38331872 ]
- Zinc finger hit domain-containing protein 1 (17 kDa, Protein from Homo sapiens)
- Actin-related protein 6 (45 kDa, Protein from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Dna methyltransferase 1-associated protein 1 (53 kDa, Protein from Homo sapiens)
- Dna (137-mer) (41 kDa, DNA from synthetic construct)
- Histone h2b type 1-c/e/f/g/i (13 kDa, Protein from Homo sapiens)
- Ruvb-like 2 (51 kDa, Protein from Homo sapiens)
- Yeats domain-containing protein 4 (26 kDa, Protein from Homo sapiens)
- Structure of nucleosome-bound srcap-c in the apo state (Complex from Homo sapiens)
- Ruvb-like 1 (50 kDa, Protein from Homo sapiens)
- Helicase srcap (343 kDa, Protein from Homo sapiens)
- Histone h2a type 1-c (14 kDa, Protein from Homo sapiens)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Dna (137-mer) (42 kDa, DNA from synthetic construct)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Actin, cytoplasmic 1, n-terminally processed (41 kDa, Protein from Homo sapiens)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Actin-like protein 6a (47 kDa, Protein from Homo sapiens)
- Vacuolar protein sorting-associated protein 72 homolog (40 kDa, Protein from Homo sapiens)
Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8x19
Q-score: 0.292
Yu J, Sui F, Gu F, Li W, Yu Z, Wang Q, He S, Wang L, Xu Y
Cell Discov (2024) 10 pp. 15-15 [ DOI: doi:10.1038/s41421-023-00640-1 Pubmed: 38331872 ]
- Zinc finger hit domain-containing protein 1 (17 kDa, Protein from Homo sapiens)
- Actin-related protein 6 (45 kDa, Protein from Homo sapiens)
- Beryllium trifluoride ion (66 Da, Ligand)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Dna methyltransferase 1-associated protein 1 (53 kDa, Protein from Homo sapiens)
- Histone h2b type 1-c/e/f/g/i (13 kDa, Protein from Homo sapiens)
- Actin, cytoplasmic 1 (41 kDa, Protein from Homo sapiens)
- Ruvb-like 2 (51 kDa, Protein from Homo sapiens)
- Yeats domain-containing protein 4 (26 kDa, Protein from Homo sapiens)
- Ruvb-like 1 (50 kDa, Protein from Homo sapiens)
- Helicase srcap (343 kDa, Protein from Homo sapiens)
- Structure of nucleosome-bound srcap-c in the adp-befx-bound state (Complex from Homo sapiens)
- Histone h2a type 1-c (14 kDa, Protein from Homo sapiens)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Dna (147-mer) (45 kDa, DNA from Homo sapiens)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Actin-like protein 6a (47 kDa, Protein from Homo sapiens)
- Dna (147-mer) (45 kDa, DNA from Homo sapiens)
- Vacuolar protein sorting-associated protein 72 homolog (40 kDa, Protein from Homo sapiens)
Structure of nucleosome-bound SRCAP-C in the ADP-bound state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8x1c
Q-score: 0.297
Yu J, Sui F, Gu F, Li W, Yu Z, Wang Q, He S, Wang L, Xu Y
Cell Discov (2024) 10 pp. 15-15 [ DOI: doi:10.1038/s41421-023-00640-1 Pubmed: 38331872 ]
- Zinc finger hit domain-containing protein 1 (17 kDa, Protein from Homo sapiens)
- Actin-related protein 6 (45 kDa, Protein from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Dna methyltransferase 1-associated protein 1 (53 kDa, Protein from Homo sapiens)
- Dna (147-mer) (45 kDa, DNA from synthetic construct)
- Histone h2b type 1-c/e/f/g/i (13 kDa, Protein from Homo sapiens)
- Actin, cytoplasmic 1 (41 kDa, Protein from Homo sapiens)
- Ruvb-like 2 (51 kDa, Protein from Homo sapiens)
- Yeats domain-containing protein 4 (26 kDa, Protein from Homo sapiens)
- Dna (147-mer) (45 kDa, DNA from synthetic construct)
- Structure of nucleosome-bound srcap-c in the adp-bound state (Complex from Homo sapiens)
- Ruvb-like 1 (50 kDa, Protein from Homo sapiens)
- Helicase srcap (343 kDa, Protein from Homo sapiens)
- Histone h2a type 1-c (14 kDa, Protein from Homo sapiens)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Actin-like protein 6a (47 kDa, Protein from Homo sapiens)
- Vacuolar protein sorting-associated protein 72 homolog (40 kDa, Protein from Homo sapiens)
Native SUV420H1 bound to 167-bp nucleosome
Resolution: 3.58 Å
EM Method: Single-particle
Fitted PDBs: 8jhg
Q-score: 0.456
Lin F, Zhang R, Shao W, Lei C, Ma M, Zhang Y, Wen Z, Li W
Cell Discov (2023) 9 pp. 120-120 [ Pubmed: 38052811 DOI: doi:10.1038/s41421-023-00620-5 ]
- Histone-lysine n-methyltransferase kmt5b (41 kDa, Protein from Homo sapiens)
- Histone h4 (9 kDa, Protein from Homo sapiens)
- Histone h2a type 1-b/e (12 kDa, Protein from Homo sapiens)
- Dna (160-mer) (49 kDa, DNA from Homo sapiens)
- Cryo-em structure of the native suv420h1 bound to 167-bp nucleosome (246 kDa, Complex from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- Dna (160-mer) (49 kDa, DNA from Homo sapiens)
- S-adenosylmethionine (398 Da, Ligand)
- Zinc ion (65 Da, Ligand)
Native SUV420H1 bound to 167-bp nucleosome
Resolution: 3.68 Å
EM Method: Single-particle
Fitted PDBs: 8jhf
Q-score: 0.433
Lin F, Zhang R, Shao W, Lei C, Ma M, Zhang Y, Wen Z, Li W
Cell Discov (2023) 9 pp. 120-120 [ Pubmed: 38052811 DOI: doi:10.1038/s41421-023-00620-5 ]
- Histone-lysine n-methyltransferase kmt5b (41 kDa, Protein from Homo sapiens)
- Histone h4 (9 kDa, Protein from Homo sapiens)
- Dna (160-mer) (49 kDa, DNA from Homo sapiens)
- Histone h2a.z (12 kDa, Protein from Homo sapiens)
- Cryo-em structure of the native suv420h1 bound to 167-bp nucleosome (246 kDa, Complex from Homo sapiens)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- Dna (160-mer) (49 kDa, DNA from Homo sapiens)
- S-adenosylmethionine (398 Da, Ligand)
- Zinc ion (65 Da, Ligand)
Structure of nucleosome-AAG complex (T-50I, post-catalytic state)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7xfj
Q-score: 0.466
Cell Discov (2023) 9 pp. 62-62 [ DOI: doi:10.1038/s41421-023-00560-0 Pubmed: 37339965 ]
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Complex (Complex)
- Dna (152-mer) (47 kDa, DNA from Xenopus laevis)
- Dna-3-methyladenine glycosylase (Complex from Homo sapiens)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Dna-3-methyladenine glycosylase (32 kDa, Protein from Homo sapiens)
- Histone h2a type 1 (14 kDa, Protein from Xenopus laevis)
- Dna (152-mer) (46 kDa, DNA from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Nucleosome (Complex from Xenopus laevis)
Structure of nucleosome-AAG complex (A-55I, post-catalytic state)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 7xnp
Q-score: 0.563
Cell Discov (2023) 9 pp. 62-62 [ DOI: doi:10.1038/s41421-023-00560-0 Pubmed: 37339965 ]
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Dna (152-mer) (47 kDa, DNA from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Dna (152-mer) (46 kDa, DNA from Xenopus laevis)
- Histone h2a type 1 (14 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Complex (Complex from Xenopus laevis)
Structure of nucleosome-DI complex (-30I, Apo state)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 7xfc
Q-score: 0.569
Cell Discov (2023) 9 pp. 62-62 [ DOI: doi:10.1038/s41421-023-00560-0 Pubmed: 37339965 ]
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Dna (152-mer) (47 kDa, DNA from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Histone h2a type 1 (14 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Dna (152-mer) (46 kDa, DNA from Xenopus laevis)
- Complex (Complex from Xenopus laevis)
Structure of nucleosome-AAG complex (A-30I, post-catalytic state)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 7xfh
Q-score: 0.515
Cell Discov (2023) 9 pp. 62-62 [ DOI: doi:10.1038/s41421-023-00560-0 Pubmed: 37339965 ]
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Complex (Complex)
- Dna-3-methyladenine glycosylase (Complex from Homo sapiens)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Dna-3-methyladenine glycosylase (32 kDa, Protein from Homo sapiens)
- Histone h2a type 1 (14 kDa, Protein from Xenopus laevis)
- Dna (152-mer) (47 kDa, DNA from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Dna (152-mer) (46 kDa, DNA from Xenopus laevis)
- Nucleosome (Complex from Xenopus laevis)
Structure of nucleosome-DI complex (-50I, Apo state)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 7xfi
Q-score: 0.525
Cell Discov (2023) 9 pp. 62-62 [ DOI: doi:10.1038/s41421-023-00560-0 Pubmed: 37339965 ]
- Dna (152-mer) (47 kDa, DNA from Xenopus laevis)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Histone h2a type 1 (14 kDa, Protein from Xenopus laevis)
- Dna (152-mer) (46 kDa, DNA from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Complex (Complex from Xenopus laevis)