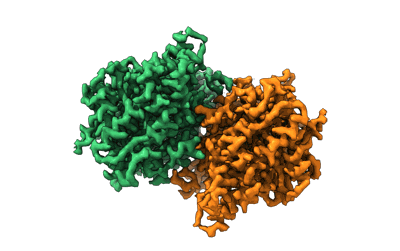
Resolution: 4.2 Å
EM Method: Helical reconstruction
Fitted PDBs: 8orh
Q-score: 0.571
Alempic JM, Bisio H, Villalta A, Santini S, Lartigue A, Schmitt A, Bugnot C, Notaro A, Belmudes L, Adrait A, Poirot O, Ptchelkine D, De Castro C, Coute Y, Abergel C
Microlife (2024) 5 pp. uqae006-uqae006 [ DOI: doi:10.1093/femsml/uqae006 Pubmed: 38659623 ]
- Putative gmc-type oxidoreductase (77 kDa, Protein from Mimivirus reunion)
- Mimivirus reunion (Virus from Mimivirus reunion)
- Flavin-adenine dinucleotide (785 Da, Ligand)
Resolution: 4.3 Å
EM Method: Helical reconstruction
Fitted PDBs: 8ors
Q-score: 0.484
Alempic JM, Bisio H, Villalta A, Santini S, Lartigue A, Schmitt A, Bugnot C, Notaro A, Belmudes L, Adrait A, Poirot O, Ptchelkine D, De Castro C, Coute Y, Abergel C
Microlife (2024) 5 pp. uqae006-uqae006 [ DOI: doi:10.1093/femsml/uqae006 Pubmed: 38659623 ]
- Putative gmc-type oxidoreductase (77 kDa, Protein from Mimivirus reunion)
- Mimivirus reunion (Virus from Mimivirus reunion)
- Flavin-adenine dinucleotide (785 Da, Ligand)
Tomogram of the Mimivirus genomic fiber
Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C
eLife (2022) 11 [ DOI: doi:10.7554/eLife.77607 Pubmed: 35900198 ]
Tomogram of the Mimivirus genomic fiber
Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C
eLife (2022) 11 [ DOI: doi:10.7554/eLife.77607 Pubmed: 35900198 ]
Tomogram of the Mimivirus genomic fiber
Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C
eLife (2022) 11 [ DOI: doi:10.7554/eLife.77607 Pubmed: 35900198 ]
Tomogram of the Mimivirus genomic fiber
Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C
eLife (2022) 11 [ DOI: doi:10.7554/eLife.77607 Pubmed: 35900198 ]
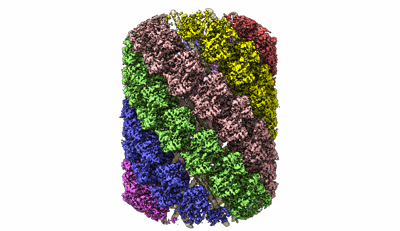
Structure of the Mimivirus genomic fibre asymmetric unit
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7ptv
Q-score: 0.579
Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C
eLife (2022) 11 [ DOI: doi:10.7554/eLife.77607 Pubmed: 35900198 ]
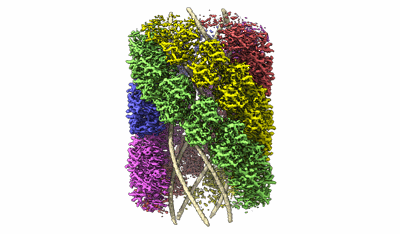
Structure of the Mimivirus genomic fibre in its compact 6-start helix form
Resolution: 4.0 Å
EM Method: Helical reconstruction
Fitted PDBs: 7yx3
Q-score: 0.458
Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C
eLife (2022) 11 [ DOI: doi:10.7554/eLife.77607 Pubmed: 35900198 ]
Structure of the Mimivirus genomic fibre in its compact 5-start helix form
Resolution: 3.7 Å
EM Method: Helical reconstruction
Fitted PDBs: 7yx4
Q-score: 0.481
Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C
eLife (2022) 11 [ DOI: doi:10.7554/eLife.77607 Pubmed: 35900198 ]
Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form
Resolution: 3.7 Å
EM Method: Helical reconstruction
Fitted PDBs: 7yx5
Q-score: 0.468
Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C
eLife (2022) 11 [ Pubmed: 35900198 DOI: doi:10.7554/eLife.77607 ]