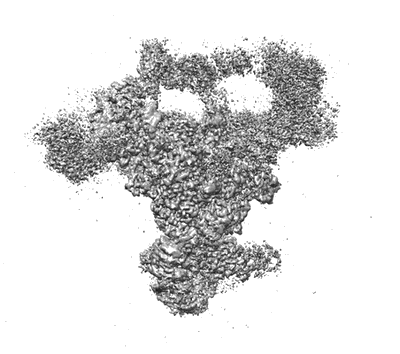
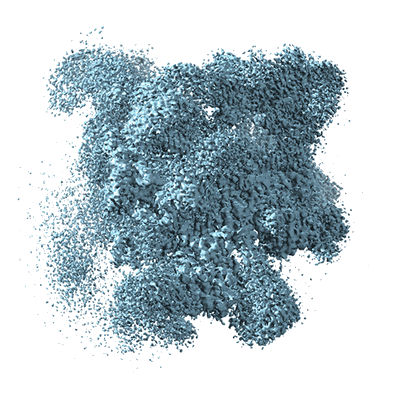
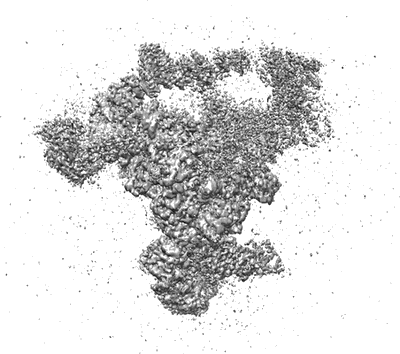Resolution: 5.9 Å
EM Method: Single-particle
Fitted PDBs: 8ch6
Q-score: 0.154
Schmitzova J, Cretu C, Dienemann C, Urlaub H, Pena V
Nature (2023) 617 pp. 842-850 [ DOI: doi:10.1038/s41586-023-06049-w Pubmed: 37165190 ]
- Splicing factor 3b subunit 4 (44 kDa, Protein from Homo sapiens)
- U5 small nuclear ribonucleoprotein 40 kda protein (39 kDa, Protein from Homo sapiens)
- Rnu2-1 (60 kDa, RNA from Homo sapiens)
- Small nuclear ribonucleoprotein e (10 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor isy1 homolog (33 kDa, Protein from Homo sapiens)
- Splicing factor 3b subunit 1 (146 kDa, Protein from Homo sapiens)
- Splicing factor 3a subunit 1 (88 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor rbm22 (46 kDa, Protein from Homo sapiens)
- Pre-mrna-processing-splicing factor 8 (273 kDa, Protein from Homo sapiens)
- G-patch domain and kow motifs-containing protein (52 kDa, Protein from Homo sapiens)
- Splicing factor 3a subunit 3 (58 kDa, Protein from Homo sapiens)
- U5 small nuclear ribonucleoprotein 200 kda helicase (244 kDa, Protein from Homo sapiens)
- Pre-mrna-processing factor 17 (65 kDa, Protein from Homo sapiens)
- Late human activated spliceosome arrested with a dominant-negative mutant of the splicing helicase aquarius (aquarius k829a) (Complex from Homo sapiens)
- Crooked neck-like protein 1 (100 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor cwc22 homolog (105 kDa, Protein from Homo sapiens)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Small nuclear ribonucleoprotein sm d3 (13 kDa, Protein from Homo sapiens)
- Small nuclear ribonucleoprotein g (8 kDa, Protein from Homo sapiens)
- Snw domain-containing protein 1 (61 kDa, Protein from Homo sapiens)
- Protein bud31 homolog (17 kDa, Protein from Homo sapiens)
- Ring-type e3 ubiquitin-protein ligase ppil2 (58 kDa, Protein from Homo sapiens)
- 116 kda u5 small nuclear ribonucleoprotein component (109 kDa, Protein from Homo sapiens)
- Small nuclear ribonucleoprotein sm d2 (13 kDa, Protein from Homo sapiens)
- Peptidyl-prolyl cis-trans isomerase-like 4 (57 kDa, Protein from Homo sapiens)
- Ring finger protein 113a (38 kDa, Protein from Homo sapiens)
- Splicing factor 3b subunit 5 (10 kDa, Protein from Homo sapiens)
- Serine/arginine repetitive matrix protein 2 (300 kDa, Protein from Homo sapiens)
- Minx-m3 (102 kDa, RNA from unidentified adenovirus)
- Peptidyl-prolyl cis-trans isomerase-like 1 (18 kDa, Protein from Homo sapiens)
- U2 small nuclear ribonucleoprotein a' (28 kDa, Protein from Homo sapiens)
- U2 small nuclear ribonucleoprotein b'' (25 kDa, Protein from Homo sapiens)
- Splicing factor 3b subunit 2 (100 kDa, Protein from Homo sapiens)
- Small nuclear ribonucleoprotein sm d1 (13 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Inositol hexakisphosphate (660 Da, Ligand)
- Small nuclear ribonucleoprotein f (9 kDa, Protein from Homo sapiens)
- Putative pre-mrna-splicing factor atp-dependent rna helicase dhx16 (119 kDa, Protein from Homo sapiens)
- Rnu6-1 (34 kDa, RNA from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Pleiotropic regulator 1 (57 kDa, Protein from Homo sapiens)
- Cell division cycle 5-like protein (92 kDa, Protein from Homo sapiens)
- Pre-mrna-processing factor 19 (55 kDa, Protein from Homo sapiens)
- Spliceosome-associated protein cwc15 homolog (26 kDa, Protein from Homo sapiens)
- Phd finger-like domain-containing protein 5a (12 kDa, Protein from Homo sapiens)
- Splicing factor 3a subunit 2 (49 kDa, Protein from Homo sapiens)
- Intron-binding protein aquarius (171 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor syf1 (100 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor spf27 (26 kDa, Protein from Homo sapiens)
- Rnu5a-1 (37 kDa, RNA from Homo sapiens)
- Peptidyl-prolyl cis-trans isomerase e (33 kDa, Protein from Homo sapiens)
- Bud13 homolog (70 kDa, Protein from Homo sapiens)
- Splicing factor 3b subunit 3 (135 kDa, Protein from Homo sapiens)
- Small nuclear ribonucleoprotein-associated proteins b and b' (24 kDa, Protein from Homo sapiens)
Yeast C complex spliceosome at 2.8 Angstrom resolution with Prp18/Slu7 bound, composite map
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 7b9v
Q-score: 0.378
Wilkinson ME, Fica SM, Galej WP, Nagai K
Mol Cell (2021) 81 pp. 1439- [ Pubmed: 33705709 DOI: doi:10.1016/j.molcel.2021.02.021 ]
- Small nuclear ribonucleoprotein f (9 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor slu7 (44 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor 18 (28 kDa, Protein from Saccharomyces cerevisiae)
- 5' exon of ubc4 mrna (15 kDa, RNA from Saccharomyces cerevisiae)
- Yeast c complex spliceosome with prp18/slu7 bound (2 MDa, Complex from Saccharomyces cerevisiae)
- Syf1 isoform 1 (100 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor slt11 (40 kDa, Protein from Saccharomyces cerevisiae)
- Unassigned structure (20 kDa, Protein from Saccharomyces cerevisiae)
- Ntc20 isoform 1 (15 kDa, Protein from Saccharomyces cerevisiae)
- U5 snrna (68 kDa, RNA from Saccharomyces cerevisiae)
- Cwc22 isoform 1 (67 kDa, Protein from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein sm d3 (11 kDa, Protein from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein g (8 kDa, Protein from Saccharomyces cerevisiae)
- U2 snrna (376 kDa, RNA from Saccharomyces cerevisiae)
- Potassium ion (39 Da, Ligand)
- U6 snrna (35 kDa, RNA from Saccharomyces cerevisiae)
- Pre-mrna-processing protein 45 (42 kDa, Protein from Saccharomyces cerevisiae)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Pre-mrna-splicing factor atp-dependent rna helicase prp16 (121 kDa, Protein from Saccharomyces cerevisiae)
- Branched intron and 3' exon of ubc4 pre-mrna (30 kDa, RNA from Saccharomyces cerevisiae)
- Hlj1_g0053790.mrna.1.cds.1 (27 kDa, Protein from Saccharomyces cerevisiae)
- Bj4_g0054360.mrna.1.cds.1 (50 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor cwc21 (15 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing helicase brr2 (246 kDa, Protein from Saccharomyces cerevisiae)
- Cdc40 isoform 1 (52 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor cwc15 (19 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-processing factor 19 (56 kDa, Protein from Saccharomyces cerevisiae)
- D-chiro inositol hexakisphosphate (660 Da, Ligand)
- Small nuclear ribonucleoprotein-associated protein b (22 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor isy1 (27 kDa, Protein from Saccharomyces cerevisiae)
- Clf1 isoform 1 (82 kDa, Protein from Saccharomyces cerevisiae)
- Bud31 isoform 1 (18 kDa, Protein from Saccharomyces cerevisiae)
- Y55_g0042700.mrna.1.cds.1 (67 kDa, Protein from Saccharomyces cerevisiae)
- Magnesium ion (24 Da, Ligand)
- Small nuclear ribonucleoprotein sm d1 (16 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor cwc2 (38 kDa, Protein from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein e (10 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor 8 (279 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor syf2 (24 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor snu114 (114 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Bj4_g0027490.mrna.1.cds.1 (12 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor cwc25 (20 kDa, Protein from Saccharomyces cerevisiae)
- Snt309 isoform 1 (20 kDa, Protein from Saccharomyces cerevisiae)
- Splicing factor yju2 (33 kDa, Protein from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein sm d2 (12 kDa, Protein from Saccharomyces cerevisiae)
Cryo-EM structure of the yeast B*-a2 complex at an average resolution of 3.2 angstrom
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 6j6g
Q-score: 0.317
Wan R, Bai R, Yan C, Lei J, Shi Y
Cell (2019) 177 pp. 339-351.e13 [ Pubmed: 30879786 DOI: doi:10.1016/j.cell.2019.02.006 ]
- U2 small nuclear ribonucleoprotein b'' (12 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-processing factor 17 (52 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor 8 (279 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor cwc2 (38 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein sm d3 (11 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein e (10 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- U5 snrna (68 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cwc21 (15 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor clf1 (82 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Small nuclear ribonucleoprotein sm d1 (16 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein g (8 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-processing factor 19 (56 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor isy1 (28 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor prp46 (50 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- U6 snrna (35 kDa, RNA from Saccharomyces cerevisiae S288c)
- Small nuclear ribonucleoprotein f (9 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor bud31 (18 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor cef1 (67 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-processing protein 45 (42 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein sm d2 (12 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Magnesium ion (24 Da, Ligand)
- Pre-mrna-splicing factor snu114 (114 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein-associated protein b (22 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Zinc ion (65 Da, Ligand)
- Pre-mrna-splicing factor syf2 (24 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- U2 snrna (376 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor snt309 (20 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Act1 pre-mrna (217 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cwc15 (19 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor syf1 (100 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor slt11 (40 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Catalytically activated spliceosome b*-a2 complex (Complex from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cwc22 (67 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Inositol hexakisphosphate (660 Da, Ligand)
- U2 small nuclear ribonucleoprotein a' (27 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 6j6h
Q-score: 0.283
Wan R, Bai R, Yan C, Lei J, Shi Y
Cell (2019) 177 pp. 339-351.e13 [ Pubmed: 30879786 DOI: doi:10.1016/j.cell.2019.02.006 ]
- U2 small nuclear ribonucleoprotein b'' (12 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-processing factor 17 (52 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor cwc2 (38 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein sm d3 (11 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein e (10 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- U5 snrna (68 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cwc21 (15 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor clf1 (82 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Catalytically activated spliceosome b*-a1 complex (Complex from Saccharomyces cerevisiae S288C)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Small nuclear ribonucleoprotein sm d1 (16 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein g (8 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-processing factor 19 (56 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor isy1 (28 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor prp46 (50 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- U6 snrna (35 kDa, RNA from Saccharomyces cerevisiae S288c)
- Small nuclear ribonucleoprotein f (9 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor bud31 (18 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor cef1 (67 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-processing protein 45 (42 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein sm d2 (12 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Magnesium ion (24 Da, Ligand)
- Pre-mrna-splicing factor 8 (279 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor snu114 (114 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein-associated protein b (22 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Zinc ion (65 Da, Ligand)
- Pre-mrna-splicing factor syf2 (24 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- U2 snrna (376 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor snt309 (20 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Act1 pre-mrna (217 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cwc15 (19 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor syf1 (100 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor slt11 (40 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor cwc22 (67 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Inositol hexakisphosphate (660 Da, Ligand)
- U2 small nuclear ribonucleoprotein a' (27 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom
Resolution: 3.86 Å
EM Method: Single-particle
Fitted PDBs: 6j6n
Q-score: 0.248
Wan R, Bai R, Yan C, Lei J, Shi Y
Cell (2019) 177 pp. 339-351.e13 [ Pubmed: 30879786 DOI: doi:10.1016/j.cell.2019.02.006 ]
- U2 small nuclear ribonucleoprotein a' (27 kDa, Protein from Saccharomyces cerevisiae S288c)
- Ubc4 pre-mrna (78 kDa, RNA from Saccharomyces cerevisiae S288c)
- Small nuclear ribonucleoprotein g (8 kDa, Protein from Saccharomyces cerevisiae S288c)
- Small nuclear ribonucleoprotein f (9 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cwc15 (19 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cwc22 (67 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-processing factor 19 (56 kDa, Protein from Saccharomyces cerevisiae S288c)
- Small nuclear ribonucleoprotein sm d3 (11 kDa, Protein from Saccharomyces cerevisiae S288c)
- U5 snrna (68 kDa, RNA from Saccharomyces cerevisiae S288c)
- Small nuclear ribonucleoprotein sm d1 (16 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cef1 (67 kDa, Protein from Saccharomyces cerevisiae S288c)
- Small nuclear ribonucleoprotein sm d2 (12 kDa, Protein from Saccharomyces cerevisiae S288c)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Pre-mrna-splicing factor slt11 (40 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor isy1 (28 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor bud31 (18 kDa, Protein from Saccharomyces cerevisiae S288c)
- U6 snrna (35 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor clf1 (82 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-processing protein 45 (42 kDa, Protein from Saccharomyces cerevisiae S288c)
- U2 small nuclear ribonucleoprotein b'' (12 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor snu114 (114 kDa, Protein from Saccharomyces cerevisiae S288c)
- Catalytically activated spliceosome b*-b1 complex (Complex from Saccharomyces cerevisiae S288C)
- Magnesium ion (24 Da, Ligand)
- Pre-mrna-splicing factor cwc21 (15 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor 8 (279 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor syf1 (100 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cwc2 (38 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor snt309 (20 kDa, Protein from Saccharomyces cerevisiae S288c)
- Inositol hexakisphosphate (660 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- U2 snrna (376 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor syf2 (24 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-processing factor 17 (52 kDa, Protein from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor prp46 (50 kDa, Protein from Saccharomyces cerevisiae S288c)
- Small nuclear ribonucleoprotein e (10 kDa, Protein from Saccharomyces cerevisiae S288c)
- Small nuclear ribonucleoprotein-associated protein b (22 kDa, Protein from Saccharomyces cerevisiae S288c)
Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 6j6q
Q-score: 0.261
Wan R, Bai R, Yan C, Lei J, Shi Y
Cell (2019) 177 pp. 339-351.e13 [ Pubmed: 30879786 DOI: doi:10.1016/j.cell.2019.02.006 ]
- U2 small nuclear ribonucleoprotein b'' (12 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Ubc4 pre-mrna (78 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-processing factor 17 (52 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor 8 (279 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor cwc2 (38 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein sm d3 (11 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein e (10 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- U5 snrna (68 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cwc21 (15 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor clf1 (82 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Small nuclear ribonucleoprotein sm d1 (16 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein g (8 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-processing factor 19 (56 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor isy1 (28 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor prp46 (50 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- U6 snrna (35 kDa, RNA from Saccharomyces cerevisiae S288c)
- Small nuclear ribonucleoprotein f (9 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor bud31 (18 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor cef1 (67 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-processing protein 45 (42 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein sm d2 (12 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Magnesium ion (24 Da, Ligand)
- Splicing factor yju2 (32 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor snu114 (114 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Small nuclear ribonucleoprotein-associated protein b (22 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Zinc ion (65 Da, Ligand)
- Pre-mrna-splicing factor syf2 (24 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- U2 snrna (376 kDa, RNA from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor snt309 (20 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor cwc15 (19 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor syf1 (100 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Pre-mrna-splicing factor slt11 (40 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Catalytically activated spliceosome b*-a2 complex (Complex from Saccharomyces cerevisiae S288c)
- Pre-mrna-splicing factor cwc22 (67 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Inositol hexakisphosphate (660 Da, Ligand)
- U2 small nuclear ribonucleoprotein a' (27 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 5yzg
Q-score: 0.205
Zhan X, Yan C, Zhang X, Lei J, Shi Y
Science (2018) 359 pp. 537-545 [ Pubmed: 29301961 DOI: doi:10.1126/science.aar6401 ]
- U5 small nuclear ribonucleoprotein 40 kda protein (39 kDa, Protein from Homo sapiens)
- Small nuclear ribonucleoprotein e (10 kDa, Protein from Homo sapiens)
- Protein casc3 (76 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor syf2 (28 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor rbm22 (46 kDa, Protein from Homo sapiens)
- Pre-mrna-processing-splicing factor 8 (273 kDa, Protein from Homo sapiens)
- Human catalytic step i spliceosome (Complex from Homo sapiens)
- Pre-mrna-splicing factor cwc25 homolog (49 kDa, Protein from Homo sapiens)
- U2 snrna (60 kDa, RNA from Homo sapiens)
- U5 small nuclear ribonucleoprotein 200 kda helicase (244 kDa, Protein from Homo sapiens)
- Pre-mrna-processing factor 17 (65 kDa, Protein from Homo sapiens)
- Crooked neck-like protein 1 (100 kDa, Protein from Homo sapiens)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Pre-mrna (88 kDa, RNA from Homo sapiens)
- Pre-mrna-splicing factor cwc22 homolog (105 kDa, Protein from Homo sapiens)
- Protein mago nashi homolog 2 (17 kDa, Protein from Homo sapiens)
- U6 snrna (34 kDa, RNA from Homo sapiens)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Small nuclear ribonucleoprotein sm d3 (13 kDa, Protein from Homo sapiens)
- Small nuclear ribonucleoprotein g (8 kDa, Protein from Homo sapiens)
- Small nuclear ribonucleoprotein-associated proteins b and b' (23 kDa, Protein from Homo sapiens)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Protein bud31 homolog (17 kDa, Protein from Homo sapiens)
- Peptidyl-prolyl cis-trans isomerase g (88 kDa, Protein from Homo sapiens)
- 116 kda u5 small nuclear ribonucleoprotein component (109 kDa, Protein from Homo sapiens)
- Small nuclear ribonucleoprotein sm d2 (13 kDa, Protein from Homo sapiens)
- Peptidylprolyl isomerase domain and wd repeat-containing protein 1 (73 kDa, Protein from Homo sapiens)
- Serine/arginine repetitive matrix protein 2 (300 kDa, Protein from Homo sapiens)
- Rna-binding protein 8a (19 kDa, Protein from Homo sapiens)
- Coiled-coil domain-containing protein 94 (37 kDa, Protein from Homo sapiens)
- U5 snrna (37 kDa, RNA from Homo sapiens)
- Peptidyl-prolyl cis-trans isomerase-like 1 (18 kDa, Protein from Homo sapiens)
- Snw domain-containing protein 1 (61 kDa, Protein from Homo sapiens)
- U2 small nuclear ribonucleoprotein a' (28 kDa, Protein from Homo sapiens)
- U2 small nuclear ribonucleoprotein b'' (25 kDa, Protein from Homo sapiens)
- Small nuclear ribonucleoprotein sm d1 (13 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor atp-dependent rna helicase prp16 (140 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Inositol hexakisphosphate (660 Da, Ligand)
- Small nuclear ribonucleoprotein f (9 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Pleiotropic regulator 1 (57 kDa, Protein from Homo sapiens)
- Eukaryotic initiation factor 4a-iii (46 kDa, Protein from Homo sapiens)
- Cell division cycle 5-like protein (92 kDa, Protein from Homo sapiens)
- Pre-mrna-processing factor 19 (55 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor isy1 homolog (35 kDa, Protein from Homo sapiens)
- Intron-binding protein aquarius (171 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor syf1 (100 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing factor spf27 (26 kDa, Protein from Homo sapiens)
- Unknow (2 kDa, Protein from Homo sapiens)
- Peptidyl-prolyl cis-trans isomerase e (33 kDa, Protein from Homo sapiens)
- Spliceosome-associated protein cwc15 homolog (26 kDa, Protein from Homo sapiens)
Structure of the yeast spliceosome immediately after branching. 3D class containing helicase module.
Resolution: 10.0 Å
EM Method: Single-particle
Fitted PDBs: 5lj5
Q-score: 0.114
Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K
Nature (2016) 537 pp. 197-201 [ DOI: doi:10.1038/nature19316 Pubmed: 27459055 ]
- Pre-mrna-splicing factor isy1 (28 kDa, Protein from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein f (9 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor bud31 (18 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor cef1 (67 kDa, Protein from Saccharomyces cerevisiae)
- Spliceosome immediately after branching (2 MDa, Complex from Saccharomyces cerevisiae)
- U2 snrna (small nuclear rna) (376 kDa, RNA from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor cwc25 (20 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor slt11 (40 kDa, Protein from Saccharomyces cerevisiae)
- U2 small nuclear ribonucleoprotein b'' (12 kDa, Protein from Saccharomyces cerevisiae)
- U2 small nuclear ribonucleoprotein a' (27 kDa, Protein from Saccharomyces cerevisiae)
- Unknown (16 kDa, Protein from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein sm d3 (11 kDa, Protein from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein g (8 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-processing protein 45 (42 kDa, Protein from Saccharomyces cerevisiae)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Pre-mrna-splicing factor atp-dependent rna helicase prp16 (121 kDa, Protein from Saccharomyces cerevisiae)
- Exon 1 (5' exon) of ubc4 pre-mrna (5 kDa, RNA from Saccharomyces cerevisiae)
- U5 snrna (small nuclear rna) (57 kDa, RNA from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor cwc21 (15 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing helicase brr2 (246 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor clf1 (82 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-processing factor 19 (56 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor snt309 (20 kDa, Protein from Saccharomyces cerevisiae)
- Intron of ubc4 pre-mrna (24 kDa, RNA from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor syf1 (100 kDa, Protein from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein-associated protein b (22 kDa, Protein from Saccharomyces cerevisiae)
- Protein cwc16 (32 kDa, Protein from Saccharomyces cerevisiae)
- Magnesium ion (24 Da, Ligand)
- U6 snrna (small nuclear rna) (35 kDa, RNA from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein sm d1 (16 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor cwc2 (38 kDa, Protein from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein e (10 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor 8 (279 kDa, Protein from Saccharomyces cerevisiae)
- Cwc22 (69 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor snu114 (114 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Cwc15 (19 kDa, Protein from Saccharomyces cerevisiae)
- Pre-mrna-splicing factor prp46 (50 kDa, Protein from Saccharomyces cerevisiae)
- Small nuclear ribonucleoprotein sm d2 (12 kDa, Protein from Saccharomyces cerevisiae)