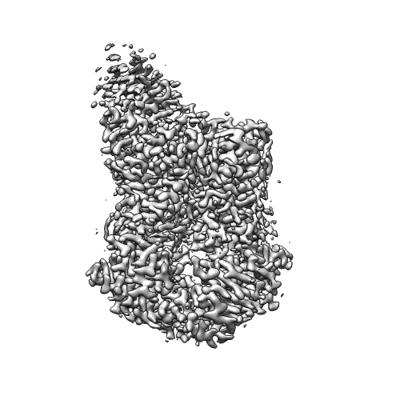
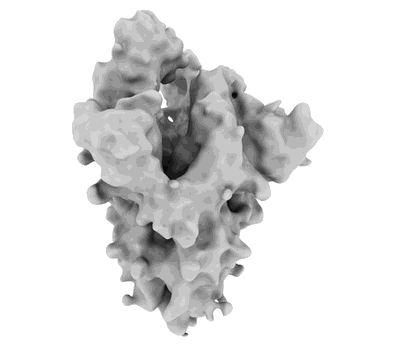
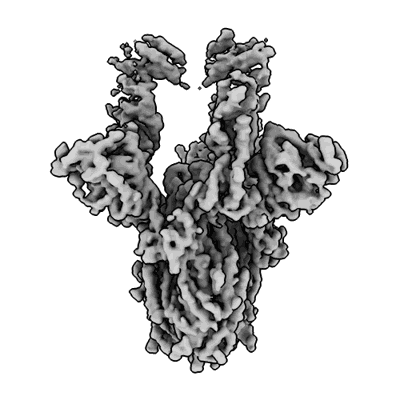PSI-LHCI supercomplex binding with 10 Lhcas from C. subellipsoidea
Resolution: 1.92 Å
EM Method: Single-particle
Fitted PDBs: 9kqp
Q-score: 0.723
Tsai PC, Kato K, Shen JR, Akita F
Biochim Biophys Acta Bioenerg (2025) 1866 pp. 149543-149543 [ Pubmed: 39947506 DOI: doi:10.1016/j.bbabio.2025.149543 ]
- Photosystem i reaction center subunit v, chloroplastic (14 kDa, Protein from Coccomyxa subellipsoidea C-169)
- 1,2-diacyl-glycerol-3-sn-phosphate (704 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (27 kDa, Protein from Coccomyxa subellipsoidea C-169)
- [2-((1-oxododecanoxy-(2-hydroxy-3-propanyl))-phosphonate-oxy)-ethyl]-trimethylammonium (440 Da, Ligand)
- Beta-carotene (536 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (29 kDa, Protein from Coccomyxa subellipsoidea C-169)
- 1,2-distearoyl-monogalactosyl-diglyceride (787 Da, Ligand)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Photosystem i p700 chlorophyll a apoprotein a2 (81 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Chlorophyll a-b binding protein, chloroplastic (29 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Photosystem i reaction center subunit ix (4 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Chlorophyll a-b binding protein, chloroplastic (26 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Photosystem i reaction center subunit ii, chloroplastic (20 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Chlorophyll b (907 Da, Ligand)
- Digalactosyl diacyl glycerol (dgdg) (949 Da, Ligand)
- Phosphatidylethanolamine (734 Da, Ligand)
- Photosystem i p700 chlorophyll a apoprotein a1 (83 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Chlorophyll a-b binding protein, chloroplastic (26 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Unknown ligand (254 Da, Ligand)
- Photosystem i psao (15 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Phylloquinone (450 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (24 kDa, Protein from Coccomyxa subellipsoidea C-169)
- (3~{e},5~{e},7~{e})-6-methyl-8-[(6~{r})-2,2,6-trimethylcyclohexyl]octa-3,5,7-trien-2-one (260 Da, Ligand)
- Green alga photosystem i lhci supercomplex binding with 10 lhcas (Complex from Coccomyxa subellipsoidea C-169)
- Photosystem i iron-sulfur center (8 kDa, Protein from Coccomyxa subellipsoidea C-169)
- 1,2-di-o-acyl-3-o-[6-deoxy-6-sulfo-alpha-d-glucopyranosyl]-sn-glycerol (795 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Photosystem i reaction center subunit iii (25 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Chlorophyll a-b binding protein, chloroplastic/lhca2 (29 kDa, Protein from Coccomyxa subellipsoidea C-169)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Water (18 Da, Ligand)
- Photosystem i reaction centre subunit vi (14 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Psi subunit v (21 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Chlorophyll a-b binding protein, chloroplastic (27 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Astaxanthin (596 Da, Ligand)
- Diacyl glycerol (625 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
- Photosystem i reaction centre subunit iv/psae (8 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Psi-k (13 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Chlorophyll a isomer (893 Da, Ligand)
- Photosystem i reaction center subunit xii (3 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Photosystem i reaction center subunit viii (3 kDa, Protein from Coccomyxa subellipsoidea C-169)
- Chlorophyll a-b binding protein, chloroplastic (24 kDa, Protein from Coccomyxa subellipsoidea C-169)
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with GABA and puerarin
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 9eqg
Q-score: 0.627
Lyu Q, Xue W, Liu R, Ma Q, Kasaragod VB, Sun S, Li Q, Chen Y, Yuan M, Yang Y, Zhang B, Nie A, Jia S, Shen C, Gao P, Rong W, Yu C, Bi Y, Zhang C, Nan F, Ning G, Rao Z, Yang X, Wang J, Wang W
Nature (2024) 634 pp. 936-943 [ Pubmed: 39261733 DOI: doi:10.1038/s41586-024-07929-5 ]
- Hexadecane (226 Da, Ligand)
- 1,2-dilauroyl-sn-glycero-3-phosphate (535 Da, Ligand)
- Hexane (86 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit gamma-2 (56 kDa, Protein from Homo sapiens)
- Gamma-aminobutyric acid receptor subunit alpha-1 (52 kDa, Protein from Homo sapiens)
- Palmitic acid (256 Da, Ligand)
- Water (18 Da, Ligand)
- (1r)-2-{[(s)-{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9z)-octadec-9-enoate (749 Da, Ligand)
- Cryoem structure of human full-length alpha1beta3gamma2l gaba(a)r in complex with gaba and puerarin (250 kDa, Complex from Homo sapiens)
- Chloride ion (35 Da, Ligand)
- Gamma-amino-butanoic acid (103 Da, Ligand)
- Decane (142 Da, Ligand)
- [(2r)-1-octadecanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8z)-icosa-5,8,11,14-tetraenoate (1 kDa, Ligand)
- Gamma-aminobutyric acid receptor subunit beta-3 (54 kDa, Protein from Homo sapiens)
- Puerarin (416 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of OSCA3.1-GDN state
Resolution: 3.11 Å
EM Method: Single-particle
Fitted PDBs: 8xw0
Q-score: 0.516
Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8ufg
Q-score: 0.527
Pahil KS, Gilman MSA, Baidin V, Clairfeuille T, Mattei P, Bieniossek C, Dey F, Muri D, Baettig R, Lobritz M, Bradley K, Kruse AC, Kahne D
Nature (2024) 625 pp. 572-577 [ Pubmed: 38172635 DOI: doi:10.1038/s41586-023-06799-7 ]
- (2~{r},4~{r},5~{r},6~{r})-2-[(2~{r},4~{r},5~{r},6~{r})-5-[(2~{s},4~{r},5~{r},6~{r})-4-[(2~{r},3~{r},4~{r},5~{s},6~{s})-3-acetamido-6-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-[(1~{r})-1,2-bis(oxidanyl)ethyl]-2-carboxy-5-oxidanyl-oxan-2-yl]oxy-6-[(1~{r})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{r},3~{s},4~{r},5~{r},6~{r})-4-[(3~{s})-3-dodecanoyloxydodecanoyl]oxy-6-[[(2~{r},3~{s},4~{r},5~{r},6~{r})-5-[[(3~{r})-3-heptanoyloxyundecanoyl]amino]-3-oxidanyl-4-[(3~{r})-3-oxidanyloctanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-5-[[(3~{s})-3-[(3~{r})-3-oxidanyldecanoyl]oxydecanoyl]amino]-3-phosphonooxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-6-[(1~{r})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid (2 kDa, Ligand)
- Lipopolysaccharide export system permease protein lptf (41 kDa, Protein from Acinetobacter baylyi ADP1)
- Inner membrane lipopolysaccharide transporter composed of lptf, lptg, and two molecules of lptb in complex with lipopolysaccharide (139 kDa, Complex from Acinetobacter baylyi ADP1)
- Lps export abc transporter permease lptg (40 kDa, Protein from Acinetobacter baylyi ADP1)
- Lipopolysaccharide export system atp-binding protein lptb (29 kDa, Protein from Acinetobacter baylyi ADP1)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8ufh
Q-score: 0.519
Pahil KS, Gilman MSA, Baidin V, Clairfeuille T, Mattei P, Bieniossek C, Dey F, Muri D, Baettig R, Lobritz M, Bradley K, Kruse AC, Kahne D
Nature (2024) 625 pp. 572-577 [ Pubmed: 38172635 DOI: doi:10.1038/s41586-023-06799-7 ]
- (2~{r},4~{r},5~{r},6~{r})-2-[(2~{r},4~{r},5~{r},6~{r})-5-[(2~{s},4~{r},5~{r},6~{r})-4-[(2~{r},3~{r},4~{r},5~{s},6~{s})-3-acetamido-6-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-[(1~{r})-1,2-bis(oxidanyl)ethyl]-2-carboxy-5-oxidanyl-oxan-2-yl]oxy-6-[(1~{r})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{r},3~{s},4~{r},5~{r},6~{r})-4-[(3~{s})-3-dodecanoyloxydodecanoyl]oxy-6-[[(2~{r},3~{s},4~{r},5~{r},6~{r})-5-[[(3~{r})-3-heptanoyloxynonanoyl]amino]-3-oxidanyl-4-[(3~{r})-3-oxidanyloctanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-5-[[(3~{s})-3-[(3~{r})-3-oxidanyldodecanoyl]oxydecanoyl]amino]-3-phosphonooxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-6-[(1~{r})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid (2 kDa, Ligand)
- Lipopolysaccharide export system permease protein lptf (41 kDa, Protein from Acinetobacter baylyi ADP1)
- (7s,10s,13s,17p)-10-(4-aminobutyl)-7-(3-aminopropyl)-17-(6-aminopyridin-3-yl)-20-chloro-13-[(1h-indol-3-yl)methyl]-12-methyl-6,7,9,10,12,13,15,16-octahydropyrido[2,3-b][1,5,8,11,14]benzothiatetraazacycloheptadecine-8,11,14(5h)-trione (797 Da, Ligand)
- Lps export abc transporter permease lptg (40 kDa, Protein from Acinetobacter baylyi ADP1)
- Lipopolysaccharide export system atp-binding protein lptb (29 kDa, Protein from Acinetobacter baylyi ADP1)
- Inner membrane lipopolysaccharide transporter composed of lptf, lptg, and two molecules of lptb in complex with lipopolysaccharide and a macrocyclic peptide (139 kDa, Complex from Acinetobacter baylyi ADP1)
Resolution: 2.785 Å
EM Method: Single-particle
Fitted PDBs: 8c29
Q-score: 0.594
Opatikova M, Semchonok DA, Kopecny D, Ilik P, Pospisil P, Ilikova I, Roudnicky P, Zeljkovic SC, Tarkowski P, Kyrilis FL, Hamdi F, Kastritis PL, Kouril R
Nat Plants (2023) 9 pp. 1359-1369 [ DOI: doi:10.1038/s41477-023-01483-0 Pubmed: 37550369 ]
- 1,2-diacyl-glycerol-3-sn-phosphate (704 Da, Ligand)
- Photosystem ii d2 protein (39 kDa, Protein from Picea abies)
- Beta-carotene (536 Da, Ligand)
- Cytochrome b559 subunit beta (4 kDa, Protein from Picea abies)
- 1,2-distearoyl-monogalactosyl-diglyceride (787 Da, Ligand)
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (32 kDa, Protein from Picea abies)
- The c2s2-type psii-lhcii supercomplex (Complex from Picea abies)
- Photosystem ii core complex (c2) (Complex from Picea abies)
- Photosystem ii psbx (12 kDa, Protein from Picea abies)
- Magnesium ion (24 Da, Ligand)
- Photosystem ii reaction center protein ycf12 (3 kDa, Protein from Picea abies)
- Chlorophyll a-b binding protein, chloroplastic (29 kDa, Protein from Picea abies)
- Palmitoleic acid (254 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- Digalactosyl diacyl glycerol (dgdg) (949 Da, Ligand)
- Bicarbonate ion (61 Da, Ligand)
- Photosystem ii reaction center protein l (4 kDa, Protein from Picea abies)
- Oxygen-evolving enhancer protein 1, chloroplastic (36 kDa, Protein from Picea abies)
- Pheophytin a (871 Da, Ligand)
- Photosystem ii reaction center protein h (7 kDa, Protein from Picea abies)
- Photosystem ii cp47 reaction center protein (56 kDa, Protein from Picea abies)
- Photosystem ii 5 kda protein, chloroplastic (14 kDa, Protein from Picea abies)
- Alpha-linolenic acid (278 Da, Ligand)
- Photosystem ii cp43 reaction center protein (51 kDa, Protein from Picea abies)
- Fe (ii) ion (55 Da, Ligand)
- Photosystem ii reaction center protein k (6 kDa, Protein from Picea abies)
- Photosystem ii reaction center protein i (4 kDa, Protein from Picea abies)
- Photosystem ii light-harvesting complex (Complex from Picea abies)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Photosystem ii reaction center protein z (6 kDa, Protein from Picea abies)
- Photosystem ii reaction center protein m (3 kDa, Protein from Picea abies)
- 1,2-di-o-acyl-3-o-[6-deoxy-6-sulfo-alpha-d-glucopyranosyl]-sn-glycerol (795 Da, Ligand)
- Cytochrome b559 subunit alpha (9 kDa, Protein from Picea abies)
- Chlorophyll a (893 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Water (18 Da, Ligand)
- Diacyl glycerol (625 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (32 kDa, Protein from Picea abies)
- Photosystem ii reaction center protein t (3 kDa, Protein from Picea abies)
- 2,3-dimethyl-5-(3,7,11,15,19,23,27,31,35-nonamethyl-2,6,10,14,18,22,26,30,34-hexatriacontanonaenyl-2,5-cyclohexadiene-1,4-dione-2,3-dimethyl-5-solanesyl-1,4-benzoquinone (749 Da, Ligand)
- (2r)-2,5,7,8-tetramethyl-2-[(4r,8r)-4,8,12-trimethyltridecyl]chroman-6-ol (430 Da, Ligand)
- Photosystem ii protein d1 (38 kDa, Protein from Picea abies)
- Psii 6.1 kda protein (15 kDa, Protein from Picea abies)
cryo-EM structure of thioredoxin glutathione reductase in complex with a non-competitive inhibitor
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8a1r
Q-score: 0.312
Petukhova VZ, Aboagye SY, Ardini M, Lullo RP, Fata F, Byrne ME, Gabriele F, Martin LM, Harding LNM, Gone V, Dangi B, Lantvit DD, Nikolic D, Ippoliti R, Effantin G, Ling WL, Johnson JJ, Thatcher GRJ, Angelucci F, Williams DL, Petukhov PA
Nat Commun (2023) 14 pp. 3737-3737 [ DOI: doi:10.1038/s41467-023-39444-y Pubmed: 37349300 ]
- Tgr in complex with an inhibitor (Complex from Schistosoma mansoni)
- Thioredoxin glutathione reductase (65 kDa, Protein from Schistosoma mansoni)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- (2~{r},3~{r},4~{s},5~{r})-2-[3-[[[(1~{r},2~{r},3~{r},5~{s})-2,6,6-trimethyl-3-bicyclo[3.1.1]heptanyl]amino]methyl]indol-1-yl]oxane-3,4,5-triol (414 Da, Ligand)
- Tgr complex with inhibitor (Complex from Schistosoma mansoni)
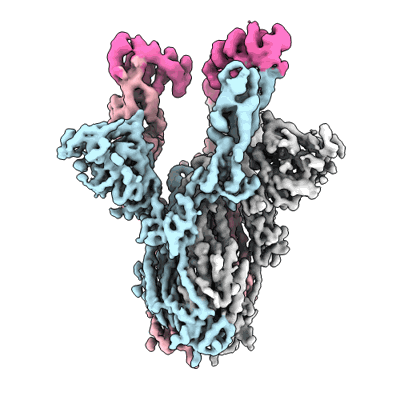
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Composite Map
Rothenberger S, Hurdiss DL, Walser M, Malvezzi F, Mayor J, Ryter S, Moreno H, Liechti N, Bosshart A, Iss C, Calabro V, Cornelius A, Hospodarsch T, Neculcea A, Looser T, Schlegel A, Fontaine S, Villemagne D, Paladino M, Schiegg D, Mangold S, Reichen C, Radom F, Kaufmann Y, Schaible D, Schlegel I, Zitt C, Sigrist G, Straumann M, Wolter J, Comby M, Sacarcelik F, Drulyte I, Lyoo H, Wang C, Li W, Du W, Binz HK, Herrup R, Lusvarghi S, Neerukonda SN, Vassell R, Wang W, Adler JM, Eschke K, Nascimento M, Abdelgawad A, Gruber AD, Bushe J, Kershaw O, Knutson CG, Balavenkatraman KK, Ramanathan K, Wyler E, Teixeira Alves LG, Lewis S, Watson R, Haeuptle MA, Zurcher A, Dawson KM, Steiner D, Weiss CD, Amstutz P, van Kuppeveld FJM, Stumpp MT, Bosch BJ, Engler O, Trimpert J
Nat Biotechnol (2022) [ Pubmed: 35864170 DOI: doi:10.1038/s41587-022-01382-3 ]
- Monovalent darpin r2 (Protein from synthetic construct)
- Sars-cov-2 spike glycoprotein (Complex from Severe acute respiratory syndrome-related coronavirus)
- Monovalent darpin r2 (Complex from Synthetic construct)
- Sars-cov-2 s 2p trimer in complex with monovalent darpin r2 (state 1) (464 kDa, Complex)
- Sars-cov-2 spike glycoprotein (Protein from Severe acute respiratory syndrome-related coronavirus)
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 2)
Rothenberger S, Hurdiss DL, Walser M, Malvezzi F, Mayor J, Ryter S, Moreno H, Liechti N, Bosshart A, Iss C, Calabro V, Cornelius A, Hospodarsch T, Neculcea A, Looser T, Schlegel A, Fontaine S, Villemagne D, Paladino M, Schiegg D, Mangold S, Reichen C, Radom F, Kaufmann Y, Schaible D, Schlegel I, Zitt C, Sigrist G, Straumann M, Wolter J, Comby M, Sacarcelik F, Drulyte I, Lyoo H, Wang C, Li W, Du W, Binz HK, Herrup R, Lusvarghi S, Neerukonda SN, Vassell R, Wang W, Adler JM, Eschke K, Nascimento M, Abdelgawad A, Gruber AD, Bushe J, Kershaw O, Knutson CG, Balavenkatraman KK, Ramanathan K, Wyler E, Teixeira Alves LG, Lewis S, Watson R, Haeuptle MA, Zurcher A, Dawson KM, Steiner D, Weiss CD, Amstutz P, van Kuppeveld FJM, Stumpp MT, Bosch BJ, Engler O, Trimpert J
Nat Biotechnol (2022) [ Pubmed: 35864170 DOI: doi:10.1038/s41587-022-01382-3 ]
- Monovalent darpin r2 (Protein from synthetic construct)
- Sars-cov-2 spike glycoprotein (Complex from Severe acute respiratory syndrome-related coronavirus)
- Sars-cov-2 s 2p trimer in complex with monovalent darpin r2 (state 2) (464 kDa, Complex)
- Monovalent darpin r2 (Complex from synthetic construct)
- Sars-cov-2 spike glycoprotein (Protein from Severe acute respiratory syndrome-related coronavirus)
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Consensus Map
Rothenberger S, Hurdiss DL, Walser M, Malvezzi F, Mayor J, Ryter S, Moreno H, Liechti N, Bosshart A, Iss C, Calabro V, Cornelius A, Hospodarsch T, Neculcea A, Looser T, Schlegel A, Fontaine S, Villemagne D, Paladino M, Schiegg D, Mangold S, Reichen C, Radom F, Kaufmann Y, Schaible D, Schlegel I, Zitt C, Sigrist G, Straumann M, Wolter J, Comby M, Sacarcelik F, Drulyte I, Lyoo H, Wang C, Li W, Du W, Binz HK, Herrup R, Lusvarghi S, Neerukonda SN, Vassell R, Wang W, Adler JM, Eschke K, Nascimento M, Abdelgawad A, Gruber AD, Bushe J, Kershaw O, Knutson CG, Balavenkatraman KK, Ramanathan K, Wyler E, Teixeira Alves LG, Lewis S, Watson R, Haeuptle MA, Zurcher A, Dawson KM, Steiner D, Weiss CD, Amstutz P, van Kuppeveld FJM, Stumpp MT, Bosch BJ, Engler O, Trimpert J
Nat Biotechnol (2022) 40 pp. 1845-1854 [ Pubmed: 35864170 DOI: doi:10.1038/s41587-022-01382-3 ]
- Monovalent darpin r2 (Protein from synthetic construct)
- Sars-cov-2 spike glycoprotein (Complex from Severe acute respiratory syndrome-related coronavirus)
- Sars-cov-2 s 2p trimer in complex with monovalent darpin r2 (464 kDa, Complex)
- Monovalent darpin r2 (Complex from synthetic construct)
- Sars-cov-2 spike glycoprotein (Protein from Severe acute respiratory syndrome coronavirus 2)