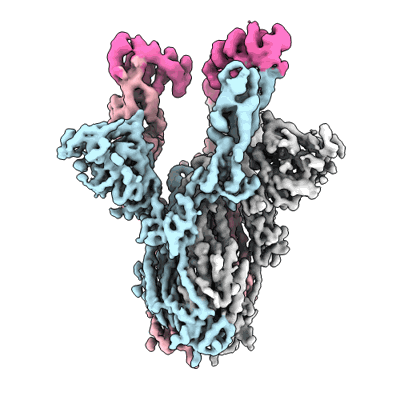
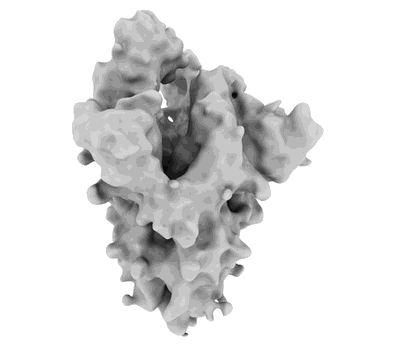
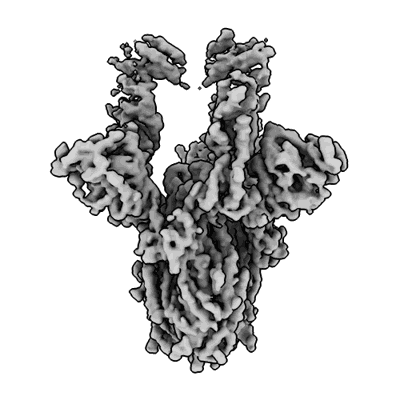Cryo-EM structure of the apo yeast Ceramide Synthase
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8qtn
Q-score: 0.513
Schafer JH, Clausmeyer L, Korner C, Esch BM, Wolf VN, Sapia J, Ahmed Y, Walter S, Vanni S, Januliene D, Moeller A, Frohlich F
Nat Struct Mol Biol (2024) [ Pubmed: 39528796 DOI: doi:10.1038/s41594-024-01415-2 ]
- Ceramide synthase subunit lip1 (15 kDa, Protein from Saccharomyces cerevisiae)
- Ceramide synthase lag1 (37 kDa, Protein from Saccharomyces cerevisiae)
- [(2s)-1-hexadecanoyloxy-3-[hydroxy-[(2s,3r,5s,6r)-2,3,4,5,6-pentahydroxycyclohexyl]oxy-phosphoryl]oxy-propan-2-yl] heptadecanoate (825 Da, Ligand)
- Ceramide synthase lac1 (36 kDa, Protein from Saccharomyces cerevisiae)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Ammonium ion (18 Da, Ligand)
- Hexacosanoic acid (396 Da, Ligand)
- Heterotetrameric complex of yeast ceramide synthase with lag1, lac1 and lip1 (140 kDa, Complex from Saccharomyces cerevisiae)
Cryo-EM structure of the FB-bound yeast Ceramide Synthase
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8qtr
Q-score: 0.464
Schafer JH, Clausmeyer L, Korner C, Esch BM, Wolf VN, Sapia J, Ahmed Y, Walter S, Vanni S, Januliene D, Moeller A, Frohlich F
Nat Struct Mol Biol (2024) [ Pubmed: 39528796 DOI: doi:10.1038/s41594-024-01415-2 ]
- Ceramide synthase lac1 (35 kDa, Protein from Saccharomyces cerevisiae)
- Macrofusine (721 Da, Ligand)
- Ceramide synthase lag1 (37 kDa, Protein from Saccharomyces cerevisiae)
- [(2s)-1-hexadecanoyloxy-3-[hydroxy-[(2s,3r,5s,6r)-2,3,4,5,6-pentahydroxycyclohexyl]oxy-phosphoryl]oxy-propan-2-yl] heptadecanoate (825 Da, Ligand)
- Ammonium ion (18 Da, Ligand)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Ceramide synthase subunit lip1 (15 kDa, Protein from Saccharomyces cerevisiae)
- Hexacosanoic acid (396 Da, Ligand)
- Heterotetrameric complex of yeast ceramide synthase with lag1, lac1 and lip1 (140 kDa, Complex from Saccharomyces cerevisiae)
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with GABA and puerarin
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 9eqg
Q-score: 0.627
Lyu Q, Xue W, Liu R, Ma Q, Kasaragod VB, Sun S, Li Q, Chen Y, Yuan M, Yang Y, Zhang B, Nie A, Jia S, Shen C, Gao P, Rong W, Yu C, Bi Y, Zhang C, Nan F, Ning G, Rao Z, Yang X, Wang J, Wang W
Nature (2024) 634 pp. 936-943 [ Pubmed: 39261733 DOI: doi:10.1038/s41586-024-07929-5 ]
- Hexadecane (226 Da, Ligand)
- 1,2-dilauroyl-sn-glycero-3-phosphate (535 Da, Ligand)
- Hexane (86 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit gamma-2 (56 kDa, Protein from Homo sapiens)
- Gamma-aminobutyric acid receptor subunit alpha-1 (52 kDa, Protein from Homo sapiens)
- Palmitic acid (256 Da, Ligand)
- Water (18 Da, Ligand)
- (1r)-2-{[(s)-{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9z)-octadec-9-enoate (749 Da, Ligand)
- Cryoem structure of human full-length alpha1beta3gamma2l gaba(a)r in complex with gaba and puerarin (250 kDa, Complex from Homo sapiens)
- Chloride ion (35 Da, Ligand)
- Gamma-amino-butanoic acid (103 Da, Ligand)
- Decane (142 Da, Ligand)
- [(2r)-1-octadecanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8z)-icosa-5,8,11,14-tetraenoate (1 kDa, Ligand)
- Gamma-aminobutyric acid receptor subunit beta-3 (54 kDa, Protein from Homo sapiens)
- Puerarin (416 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8gz2
Q-score: 0.422
Li Y, Yuan T, Huang B, Zhou F, Peng C, Li X, Qiu Y, Yang B, Zhao Y, Huang Z, Jiang D
Nat Commun (2023) 14 pp. 1030-1030 [ Pubmed: 36823201 DOI: doi:10.1038/s41467-023-36766-9 ]
- Sodium channel protein type 8 subunit alpha (225 kDa, Protein from Homo sapiens)
- Sodium channel subunit beta-2 (24 kDa, Protein from Homo sapiens)
- Structure of human voltage-gated sodium channel nav1.6 in complex with auxiliary beta subunits (Complex from Homo sapiens)
- Sodium channel subunit beta-1 (24 kDa, Protein from Homo sapiens)
- (1r,2s,3s,4r,5r,9s,11s,12s,14r)-7-amino-2,4,12-trihydroxy-2-(hydroxymethyl)-10,13,15-trioxa-6,8-diazapentacyclo[7.4.1.1~3,12~.0~5,11~.0~5,14~]pentadec-7-en-8-ium (non-preferred name) (301 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Phophorylated Drs2p-Cdc50p in a PS and ATP-bound E2P state
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7pem
Q-score: 0.516
Wang Y, Lyons JA, Timcenko M, Kummerer F, de Groot BL, Nissen P, Gapsys V, Lindorff-Larsen K
To Be Published
- Binary complex of drs2p-cdc50p with the regulatory lipid pi4p and transport substrate ps (180 kDa, Complex from Saccharomyces cerevisiae)
- Cell division control protein 50 (45 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Probable phospholipid-transporting atpase drs2 (154 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- O-[(r)-[(2s)-2-(hexadecanoyloxy)-3-(octadecanoyloxy)propoxy](hydroxy)phosphoryl]-d-serine (764 Da, Ligand)
- (2r)-1-{[(r)-hydroxy{[(1r,2r,3r,4r,5s,6r)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5z,8z,11z,14z)-icosa-5,8,11,14-tetraenoate (967 Da, Ligand)
- Water (18 Da, Ligand)
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Composite Map
Rothenberger S, Hurdiss DL, Walser M, Malvezzi F, Mayor J, Ryter S, Moreno H, Liechti N, Bosshart A, Iss C, Calabro V, Cornelius A, Hospodarsch T, Neculcea A, Looser T, Schlegel A, Fontaine S, Villemagne D, Paladino M, Schiegg D, Mangold S, Reichen C, Radom F, Kaufmann Y, Schaible D, Schlegel I, Zitt C, Sigrist G, Straumann M, Wolter J, Comby M, Sacarcelik F, Drulyte I, Lyoo H, Wang C, Li W, Du W, Binz HK, Herrup R, Lusvarghi S, Neerukonda SN, Vassell R, Wang W, Adler JM, Eschke K, Nascimento M, Abdelgawad A, Gruber AD, Bushe J, Kershaw O, Knutson CG, Balavenkatraman KK, Ramanathan K, Wyler E, Teixeira Alves LG, Lewis S, Watson R, Haeuptle MA, Zurcher A, Dawson KM, Steiner D, Weiss CD, Amstutz P, van Kuppeveld FJM, Stumpp MT, Bosch BJ, Engler O, Trimpert J
Nat Biotechnol (2022) [ Pubmed: 35864170 DOI: doi:10.1038/s41587-022-01382-3 ]
- Monovalent darpin r2 (Protein from synthetic construct)
- Sars-cov-2 spike glycoprotein (Complex from Severe acute respiratory syndrome-related coronavirus)
- Monovalent darpin r2 (Complex from Synthetic construct)
- Sars-cov-2 s 2p trimer in complex with monovalent darpin r2 (state 1) (464 kDa, Complex)
- Sars-cov-2 spike glycoprotein (Protein from Severe acute respiratory syndrome-related coronavirus)
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 2)
Rothenberger S, Hurdiss DL, Walser M, Malvezzi F, Mayor J, Ryter S, Moreno H, Liechti N, Bosshart A, Iss C, Calabro V, Cornelius A, Hospodarsch T, Neculcea A, Looser T, Schlegel A, Fontaine S, Villemagne D, Paladino M, Schiegg D, Mangold S, Reichen C, Radom F, Kaufmann Y, Schaible D, Schlegel I, Zitt C, Sigrist G, Straumann M, Wolter J, Comby M, Sacarcelik F, Drulyte I, Lyoo H, Wang C, Li W, Du W, Binz HK, Herrup R, Lusvarghi S, Neerukonda SN, Vassell R, Wang W, Adler JM, Eschke K, Nascimento M, Abdelgawad A, Gruber AD, Bushe J, Kershaw O, Knutson CG, Balavenkatraman KK, Ramanathan K, Wyler E, Teixeira Alves LG, Lewis S, Watson R, Haeuptle MA, Zurcher A, Dawson KM, Steiner D, Weiss CD, Amstutz P, van Kuppeveld FJM, Stumpp MT, Bosch BJ, Engler O, Trimpert J
Nat Biotechnol (2022) [ Pubmed: 35864170 DOI: doi:10.1038/s41587-022-01382-3 ]
- Monovalent darpin r2 (Protein from synthetic construct)
- Sars-cov-2 spike glycoprotein (Complex from Severe acute respiratory syndrome-related coronavirus)
- Sars-cov-2 s 2p trimer in complex with monovalent darpin r2 (state 2) (464 kDa, Complex)
- Monovalent darpin r2 (Complex from synthetic construct)
- Sars-cov-2 spike glycoprotein (Protein from Severe acute respiratory syndrome-related coronavirus)
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Consensus Map
Rothenberger S, Hurdiss DL, Walser M, Malvezzi F, Mayor J, Ryter S, Moreno H, Liechti N, Bosshart A, Iss C, Calabro V, Cornelius A, Hospodarsch T, Neculcea A, Looser T, Schlegel A, Fontaine S, Villemagne D, Paladino M, Schiegg D, Mangold S, Reichen C, Radom F, Kaufmann Y, Schaible D, Schlegel I, Zitt C, Sigrist G, Straumann M, Wolter J, Comby M, Sacarcelik F, Drulyte I, Lyoo H, Wang C, Li W, Du W, Binz HK, Herrup R, Lusvarghi S, Neerukonda SN, Vassell R, Wang W, Adler JM, Eschke K, Nascimento M, Abdelgawad A, Gruber AD, Bushe J, Kershaw O, Knutson CG, Balavenkatraman KK, Ramanathan K, Wyler E, Teixeira Alves LG, Lewis S, Watson R, Haeuptle MA, Zurcher A, Dawson KM, Steiner D, Weiss CD, Amstutz P, van Kuppeveld FJM, Stumpp MT, Bosch BJ, Engler O, Trimpert J
Nat Biotechnol (2022) 40 pp. 1845-1854 [ Pubmed: 35864170 DOI: doi:10.1038/s41587-022-01382-3 ]
- Monovalent darpin r2 (Protein from synthetic construct)
- Sars-cov-2 spike glycoprotein (Complex from Severe acute respiratory syndrome-related coronavirus)
- Sars-cov-2 s 2p trimer in complex with monovalent darpin r2 (464 kDa, Complex)
- Monovalent darpin r2 (Complex from synthetic construct)
- Sars-cov-2 spike glycoprotein (Protein from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Focused Refinement
Rothenberger S, Hurdiss DL, Walser M, Malvezzi F, Mayor J, Ryter S, Moreno H, Liechti N, Bosshart A, Iss C, Calabro V, Cornelius A, Hospodarsch T, Neculcea A, Looser T, Schlegel A, Fontaine S, Villemagne D, Paladino M, Schiegg D, Mangold S, Reichen C, Radom F, Kaufmann Y, Schaible D, Schlegel I, Zitt C, Sigrist G, Straumann M, Wolter J, Comby M, Sacarcelik F, Drulyte I, Lyoo H, Wang C, Li W, Du W, Binz HK, Herrup R, Lusvarghi S, Neerukonda SN, Vassell R, Wang W, Adler JM, Eschke K, Nascimento M, Abdelgawad A, Gruber AD, Bushe J, Kershaw O, Knutson CG, Balavenkatraman KK, Ramanathan K, Wyler E, Teixeira Alves LG, Lewis S, Watson R, Haeuptle MA, Zurcher A, Dawson KM, Steiner D, Weiss CD, Amstutz P, van Kuppeveld FJM, Stumpp MT, Bosch BJ, Engler O, Trimpert J
Nat Biotechnol (2022) 40 pp. 1845-1854 [ Pubmed: 35864170 DOI: doi:10.1038/s41587-022-01382-3 ]
- Monovalent darpin r2 (Protein from synthetic construct)
- Sars-cov-2 spike glycoprotein (Complex from Severe acute respiratory syndrome-related coronavirus)
- Sars-cov-2 s 2p trimer in complex with monovalent darpin r2 (464 kDa, Complex)
- Monovalent darpin r2 (Complex from synthetic construct)
- Sars-cov-2 spike glycoprotein (Protein from Severe acute respiratory syndrome coronavirus 2)