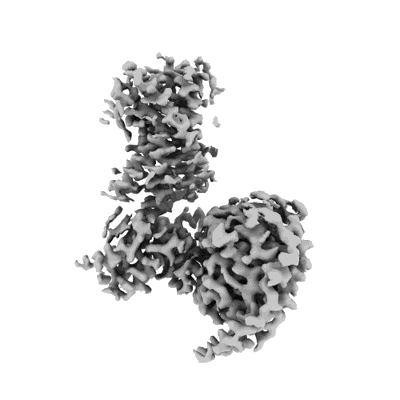
NDH-PSI-LHCI supercomplex from S. oleracea: composite map
Resolution: 3.19 Å
EM Method: Single-particle
Fitted PDBs: 9grx
Q-score: 0.541
Introini B, Hahn A, Kuhlbrandt W
Nat Struct Mol Biol (2025) [ DOI: doi:10.1038/s41594-024-01478-1 Pubmed: 39856350 ]
- Photosystem i reaction center subunit xi, chloroplastic (16 kDa, Protein from Spinacia oleracea)
- Ndh-psi-lhci supercomplex from spinacia oleracea (Complex from Spinacia oleracea)
- Photosystem i reaction center subunit viii (3 kDa, Protein from Spinacia oleracea)
- Chlorophyll a-b binding protein, chloroplastic (21 kDa, Protein from Spinacia oleracea)
- Nad(p)h-quinone oxidoreductase subunit 5, chloroplastic (84 kDa, Protein from Spinacia oleracea)
- Nad(p)h-quinone oxidoreductase subunit 6, chloroplastic (19 kDa, Protein from Spinacia oleracea)
- Beta-carotene (536 Da, Ligand)
- 4-trans-(4-trans-propylcyclohexyl)-cyclohexyl alpha-maltoside (548 Da, Ligand)
- 1,2-distearoyl-monogalactosyl-diglyceride (787 Da, Ligand)
- Nad(p)h-quinone oxidoreductase subunit 1, chloroplastic (38 kDa, Protein from Spinacia oleracea)
- Photosynthetic ndh subunit of lumenal location 3, chloroplastic (16 kDa, Protein from Spinacia oleracea)
- Nad(p)h-quinone oxidoreductase chain 4, chloroplastic (55 kDa, Protein from Spinacia oleracea)
- 5-[(2e,6e,10e,14e,18e,22e)-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18,22,26-heptaenyl]-2,3-dimethylbenzo-1,4-quinone (612 Da, Ligand)
- Nad(p)h-quinone oxidoreductase subunit k, chloroplastic (22 kDa, Protein from Spinacia oleracea)
- Psi-k (13 kDa, Protein from Spinacia oleracea)
- Nad(p)h-quinone oxidoreductase subunit n, chloroplastic (18 kDa, Protein from Spinacia oleracea)
- Nad(p)h-quinone oxidoreductase subunit i, chloroplastic (19 kDa, Protein from Spinacia oleracea)
- Chlorophyll a-b binding protein, chloroplastic (22 kDa, Protein from Spinacia oleracea)
- Photosystem i reaction center subunit iv, chloroplastic (7 kDa, Protein from Spinacia oleracea)
- Peptidyl-prolyl cis-trans isomerase (18 kDa, Protein from Spinacia oleracea)
- Digalactosyl diacyl glycerol (dgdg) (949 Da, Ligand)
- Photosystem i reaction center subunit ix (4 kDa, Protein from Spinacia oleracea)
- Photosystem i iron-sulfur center (9 kDa, Protein from Spinacia oleracea)
- Chlorophyll b (907 Da, Ligand)
- Nad(p)h-quinone oxidoreductase subunit j, chloroplastic (18 kDa, Protein from Spinacia oleracea)
- Photosynthetic ndh subunit of lumenal location 2, chloroplastic (14 kDa, Protein from Spinacia oleracea)
- Photosystem i reaction center subunit ii, chloroplastic (16 kDa, Protein from Spinacia oleracea)
- Photosynthetic ndh subunit of subcomplex b 3, chloroplastic (15 kDa, Protein from Spinacia oleracea)
- Photosynthetic ndh subunit of subcomplex b 2, chloroplastic (40 kDa, Protein from Spinacia oleracea)
- Peptidylprolyl isomerase (15 kDa, Protein from Spinacia oleracea)
- Photosystem i p700 chlorophyll a apoprotein a1 (82 kDa, Protein from Spinacia oleracea)
- Photosynthetic ndh subunit of subcomplex b 5, chloroplastic (17 kDa, Protein from Spinacia oleracea)
- Photosystem i reaction center subunit v, chloroplastic (10 kDa, Protein from Spinacia oleracea)
- Photosystem i reaction center subunit vi, chloroplastic (10 kDa, Protein from Spinacia oleracea)
- (1s)-2-{[{[(2r)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(palmitoyloxy)methyl]ethyl stearate (751 Da, Ligand)
- Nad(p)h-quinone oxidoreductase subunit h, chloroplastic (45 kDa, Protein from Spinacia oleracea)
- Phylloquinone (450 Da, Ligand)
- Photosynthetic ndh subunit of subcomplex b 1, chloroplastic (44 kDa, Protein from Spinacia oleracea)
- Photosystem i p700 chlorophyll a apoprotein a2 (82 kDa, Protein from Spinacia oleracea)
- Nad(p)h-quinone oxidoreductase subunit 3, chloroplastic (13 kDa, Protein from Spinacia oleracea)
- Nad(p)h-quinone oxidoreductase subunit o, chloroplastic (11 kDa, Protein from Spinacia oleracea)
- Chlorophyll a isomer (893 Da, Ligand)
- 1,2-di-o-acyl-3-o-[6-deoxy-6-sulfo-alpha-d-glucopyranosyl]-sn-glycerol (795 Da, Ligand)
- Nad(p)h-quinone oxidoreductase subunit 4l, chloroplastic (11 kDa, Protein from Spinacia oleracea)
- Chlorophyll a (893 Da, Ligand)
- Nad(p)h-quinone oxidoreductase subunit u, chloroplastic (27 kDa, Protein from Spinacia oleracea)
- Photosystem i reaction center subunit iii, chloroplastic (17 kDa, Protein from Spinacia oleracea)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Photosynthetic ndh subunit of subcomplex b 4, chloroplastic (10 kDa, Protein from Spinacia oleracea)
- Chlorophyll a-b binding protein, chloroplastic (24 kDa, Protein from Spinacia oleracea)
- Iron/sulfur cluster (351 Da, Ligand)
- Nad(p)h-quinone oxidoreductase subunit 2 a, chloroplastic (54 kDa, Protein from Spinacia oleracea)
- Nad(p)h-quinone oxidoreductase subunit m, chloroplastic (16 kDa, Protein from Spinacia oleracea)
- Nad(p)h-quinone oxidoreductase subunit l, chloroplastic (13 kDa, Protein from Spinacia oleracea)
- Photosynthetic ndh subunit of lumenal location 1, chloroplastic (18 kDa, Protein from Spinacia oleracea)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
Curved structure of mPIEZO1-S2472E
Liu S, Yang X, Chen X, Zhang X, Jiang J, Yuan J, Liu W, Wang L, Zhou H, Wu K, Tian B, Li X, Xiao B
Neuron (2025) 113 pp. 590- [ Pubmed: 39719701 DOI: doi:10.1016/j.neuron.2024.11.020 ]
- S2472e-curved structure (Complex from Mus musculus)
- Piezo-type mechanosensitive ion channel component 1 (292 kDa, Protein from Mus musculus)
- O-[(r)-{[(2r)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-l-serine (792 Da, Ligand)
- (9r,11s)-9-({[(1s)-1-hydroxyhexadecyl]oxy}methyl)-2,2-dimethyl-5,7,10-trioxa-2lambda~5~-aza-6lambda~5~-phosphaoctacosane-6,6,11-triol (767 Da, Ligand)
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (744 Da, Ligand)
Flattened structure of mPIEZO1-S2472E
Liu S, Yang X, Chen X, Zhang X, Jiang J, Yuan J, Liu W, Wang L, Zhou H, Wu K, Tian B, Li X, Xiao B
Neuron (2025) 113 pp. 590- [ Pubmed: 39719701 DOI: doi:10.1016/j.neuron.2024.11.020 ]
- Piezo-type mechanosensitive ion channel component 1 (292 kDa, Protein from Mus musculus)
- O-[(r)-{[(2r)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-l-serine (792 Da, Ligand)
- (9r,11s)-9-({[(1s)-1-hydroxyhexadecyl]oxy}methyl)-2,2-dimethyl-5,7,10-trioxa-2lambda~5~-aza-6lambda~5~-phosphaoctacosane-6,6,11-triol (767 Da, Ligand)
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (744 Da, Ligand)
- S2472e-intermediate structure (Complex from Mus musculus)
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with GABA and puerarin
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 9eqg
Q-score: 0.627
Lyu Q, Xue W, Liu R, Ma Q, Kasaragod VB, Sun S, Li Q, Chen Y, Yuan M, Yang Y, Zhang B, Nie A, Jia S, Shen C, Gao P, Rong W, Yu C, Bi Y, Zhang C, Nan F, Ning G, Rao Z, Yang X, Wang J, Wang W
Nature (2024) 634 pp. 936-943 [ Pubmed: 39261733 DOI: doi:10.1038/s41586-024-07929-5 ]
- Hexadecane (226 Da, Ligand)
- 1,2-dilauroyl-sn-glycero-3-phosphate (535 Da, Ligand)
- Hexane (86 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit gamma-2 (56 kDa, Protein from Homo sapiens)
- Gamma-aminobutyric acid receptor subunit alpha-1 (52 kDa, Protein from Homo sapiens)
- Palmitic acid (256 Da, Ligand)
- Water (18 Da, Ligand)
- (1r)-2-{[(s)-{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9z)-octadec-9-enoate (749 Da, Ligand)
- Cryoem structure of human full-length alpha1beta3gamma2l gaba(a)r in complex with gaba and puerarin (250 kDa, Complex from Homo sapiens)
- Chloride ion (35 Da, Ligand)
- Gamma-amino-butanoic acid (103 Da, Ligand)
- Decane (142 Da, Ligand)
- [(2r)-1-octadecanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8z)-icosa-5,8,11,14-tetraenoate (1 kDa, Ligand)
- Gamma-aminobutyric acid receptor subunit beta-3 (54 kDa, Protein from Homo sapiens)
- Puerarin (416 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Structure of TbAQP2 in complex with anti-trypanosomatid drug melarsoprol
Structure of NET-Nisoxetine in outward-open state
Resolution: 2.89 Å
EM Method: Single-particle
Fitted PDBs: 8yr2
Q-score: 0.47
Zhang H, Yin YL, Dai A, Zhang T, Zhang C, Wu C, Hu W, He X, Pan B, Jin S, Yuan Q, Wang MW, Yang D, Xu HE, Jiang Y
Nature (2024) 630 pp. 247-254 [ Pubmed: 38750358 DOI: doi:10.1038/s41586-024-07437-6 ]
- Nisoxetine (271 Da, Ligand)
- Water (18 Da, Ligand)
- [(2r)-2-octanoyloxy-3-[oxidanyl-[(2r,3r,5s,6r)-2,3,4,5,6-pentakis(oxidanyl)cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate (586 Da, Ligand)
- Sodium ion (22 Da, Ligand)
- Sodium-dependent noradrenaline transporter (69 kDa, Protein from Homo sapiens)
- Chloride ion (35 Da, Ligand)
- Cholesterol (386 Da, Ligand)
- Sodium-dependent noradrenaline transporter (Complex from Homo sapiens)
- [(2r)-2-octanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate (746 Da, Ligand)
Cryo-EM structure of OSCA3.1-GDN state
Resolution: 3.11 Å
EM Method: Single-particle
Fitted PDBs: 8xw0
Q-score: 0.516
cryo-EM structure of thioredoxin glutathione reductase in complex with a non-competitive inhibitor
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8a1r
Q-score: 0.312
Petukhova VZ, Aboagye SY, Ardini M, Lullo RP, Fata F, Byrne ME, Gabriele F, Martin LM, Harding LNM, Gone V, Dangi B, Lantvit DD, Nikolic D, Ippoliti R, Effantin G, Ling WL, Johnson JJ, Thatcher GRJ, Angelucci F, Williams DL, Petukhov PA
Nat Commun (2023) 14 pp. 3737-3737 [ DOI: doi:10.1038/s41467-023-39444-y Pubmed: 37349300 ]
- Tgr in complex with an inhibitor (Complex from Schistosoma mansoni)
- Thioredoxin glutathione reductase (65 kDa, Protein from Schistosoma mansoni)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- (2~{r},3~{r},4~{s},5~{r})-2-[3-[[[(1~{r},2~{r},3~{r},5~{s})-2,6,6-trimethyl-3-bicyclo[3.1.1]heptanyl]amino]methyl]indol-1-yl]oxane-3,4,5-triol (414 Da, Ligand)
- Tgr complex with inhibitor (Complex from Schistosoma mansoni)
Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGs
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7wu3
Q-score: 0.501
Qu X, Qiu N, Wang M, Zhang B, Du J, Zhong Z, Xu W, Chu X, Ma L, Yi C, Han S, Shui W, Zhao Q, Wu B
Nature (2022) 604 pp. 779-785 [ DOI: doi:10.1038/s41586-022-04580-w Pubmed: 35418679 ]
- Nanobody nb35 (17 kDa, Protein from Lama glama)
- Nanobody nb35 (Complex from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(s) subunit alpha isoforms short (28 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (38 kDa, Protein from Homo sapiens)
- Adhesion g-protein coupled receptor f1 (74 kDa, Protein from Homo sapiens)
- [(2~{r})-2-oxidanyl-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propyl] hexadecanoate (496 Da, Ligand)
- The adhesion gpcr adgrf1 in complex with minigs (Complex from Homo sapiens)
Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGi
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7wu4
Q-score: 0.428
Qu X, Qiu N, Wang M, Zhang B, Du J, Zhong Z, Xu W, Chu X, Ma L, Yi C, Han S, Shui W, Zhao Q, Wu B
Nature (2022) 604 pp. 779-785 [ DOI: doi:10.1038/s41586-022-04580-w Pubmed: 35418679 ]
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (27 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- The adhesion gpcr adgrf1 in complex with minigi (Complex from Homo sapiens)
- Adhesion g-protein coupled receptor f1 (74 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (38 kDa, Protein from Homo sapiens)
- [(2~{r})-2-oxidanyl-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propyl] hexadecanoate (496 Da, Ligand)
- Cholesterol (386 Da, Ligand)