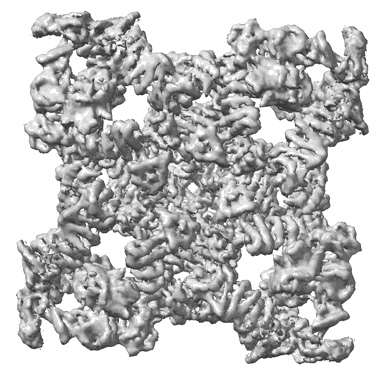
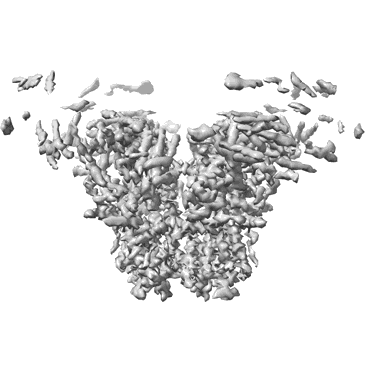
Resolution: 3.49 Å
EM Method: Single-particle
Fitted PDBs: 8sen
Q-score: 0.338
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S
Structure (2023) 31 pp. 790-800.e4 [ DOI: doi:10.1016/j.str.2023.04.009 Pubmed: 37192614 ]
- Ryr1 in complex with fkbp12.6 (Complex from Oryctolagus cuniculus)
- Ryanodine receptor 1 (565 kDa, Protein from Oryctolagus cuniculus)
- Glutathione s-transferase class-mu 26 kda isozyme,peptidyl-prolyl cis-trans isomerase fkbp1b (40 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
Cryo-EM Structure of RyR1 + ATP-gamma-S
Resolution: 3.92 Å
EM Method: Single-particle
Fitted PDBs: 8seo
Q-score: 0.253
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S
Structure (2023) 31 pp. 790-800.e4 [ DOI: doi:10.1016/j.str.2023.04.009 Pubmed: 37192614 ]
- Phosphothiophosphoric acid-adenylate ester (523 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Ryr1 in complex with fkbp12.6 (Complex from Oryctolagus cuniculus)
- Ryanodine receptor 1 (565 kDa, Protein from Oryctolagus cuniculus)
- Glutathione s-transferase class-mu 26 kda isozyme,peptidyl-prolyl cis-trans isomerase fkbp1b (40 kDa, Protein from Homo sapiens)
Cryo-EM Structure of RyR1 + ADP
Resolution: 3.57 Å
EM Method: Single-particle
Fitted PDBs: 8sep
Q-score: 0.289
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S
Structure (2023) 31 pp. 790-800.e4 [ DOI: doi:10.1016/j.str.2023.04.009 Pubmed: 37192614 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Ryr1 in complex with fkbp12.6 (Complex from Oryctolagus cuniculus)
- Ryanodine receptor 1 (565 kDa, Protein from Oryctolagus cuniculus)
- Glutathione s-transferase class-mu 26 kda isozyme,peptidyl-prolyl cis-trans isomerase fkbp1b (40 kDa, Protein from Homo sapiens)
Cryo-EM Structure of RyR1 + AMP
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8seq
Q-score: 0.322
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S
Structure (2023) 31 pp. 790-800.e4 [ DOI: doi:10.1016/j.str.2023.04.009 Pubmed: 37192614 ]
- Adenosine monophosphate (347 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Ryr1 in complex with fkbp12.6 (Complex from Oryctolagus cuniculus)
- Ryanodine receptor 1 (565 kDa, Protein from Oryctolagus cuniculus)
- Glutathione s-transferase class-mu 26 kda isozyme,peptidyl-prolyl cis-trans isomerase fkbp1b (40 kDa, Protein from Homo sapiens)
Cryo-EM Structure of RyR1 + Adenosine
Resolution: 3.42 Å
EM Method: Single-particle
Fitted PDBs: 8ser
Q-score: 0.353
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S
Structure (2023) 31 pp. 790-800.e4 [ DOI: doi:10.1016/j.str.2023.04.009 Pubmed: 37192614 ]
- Zinc ion (65 Da, Ligand)
- Ryr1 in complex with fkbp12.6 (Complex from Oryctolagus cuniculus)
- Ryanodine receptor 1 (565 kDa, Protein from Oryctolagus cuniculus)
- Adenosine (267 Da, Ligand)
- Glutathione s-transferase class-mu 26 kda isozyme,peptidyl-prolyl cis-trans isomerase fkbp1b (40 kDa, Protein from Homo sapiens)
Cryo-EM Structure of RyR1 + Adenine
Resolution: 3.98 Å
EM Method: Single-particle
Fitted PDBs: 8ses
Q-score: 0.284
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S
Structure (2023) 31 pp. 790-800.e4 [ DOI: doi:10.1016/j.str.2023.04.009 Pubmed: 37192614 ]
- Adenine (135 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Ryr1 in complex with fkbp12.6 (Complex from Oryctolagus cuniculus)
- Ryanodine receptor 1 (565 kDa, Protein from Oryctolagus cuniculus)
- Glutathione s-transferase class-mu 26 kda isozyme,peptidyl-prolyl cis-trans isomerase fkbp1b (40 kDa, Protein from Homo sapiens)
Cryo-EM Structure of RyR1 + cAMP
Resolution: 3.42 Å
EM Method: Single-particle
Fitted PDBs: 8set
Q-score: 0.309
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S
Structure (2023) 31 pp. 790-800.e4 [ DOI: doi:10.1016/j.str.2023.04.009 Pubmed: 37192614 ]
- Adenosine-3',5'-cyclic-monophosphate (329 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Ryr1 in complex with fkbp12.6 (Complex from Oryctolagus cuniculus)
- Ryanodine receptor 1 (565 kDa, Protein from Oryctolagus cuniculus)
- Glutathione s-transferase class-mu 26 kda isozyme,peptidyl-prolyl cis-trans isomerase fkbp1b (40 kDa, Protein from Homo sapiens)
Cryo-EM Structure of RyR1 (Local Refinement of TMD)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8seu
Q-score: 0.549
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S
Structure (2023) 31 pp. 790-800.e4 [ DOI: doi:10.1016/j.str.2023.04.009 Pubmed: 37192614 ]
- Ryr1 in complex with fkbp12.6 (Complex from Oryctolagus cuniculus)
- Ryanodine receptor 1 (565 kDa, Protein from Oryctolagus cuniculus)
- Zinc ion (65 Da, Ligand)
Cryo-EM Structure of RyR1 + ATP-gamma-S (Local Refinement of TMD)
Resolution: 3.17 Å
EM Method: Single-particle
Fitted PDBs: 8sev
Q-score: 0.499
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S
Structure (2023) 31 pp. 790-800.e4 [ DOI: doi:10.1016/j.str.2023.04.009 Pubmed: 37192614 ]
- Ryr1 in complex with fkbp12.6 (Complex from Oryctolagus cuniculus)
- Ryanodine receptor 1 (565 kDa, Protein from Oryctolagus cuniculus)
- Phosphothiophosphoric acid-adenylate ester (523 Da, Ligand)
- Zinc ion (65 Da, Ligand)
Cryo-EM Structure of RyR1 + ADP (Local Refinement of TMD)
Resolution: 2.89 Å
EM Method: Single-particle
Fitted PDBs: 8sew
Q-score: 0.549
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S
Structure (2023) 31 pp. 790-800.e4 [ DOI: doi:10.1016/j.str.2023.04.009 Pubmed: 37192614 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Ryr1 in complex with fkbp12.6 (Complex from Oryctolagus cuniculus)
- Ryanodine receptor 1 (565 kDa, Protein from Oryctolagus cuniculus)
- Zinc ion (65 Da, Ligand)