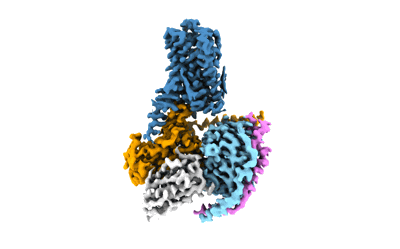
Cryo-EM structure of the RO5256390-TAAR1 complex
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8w8a
Q-score: 0.551
Liu H, Zheng Y, Wang Y, Wang Y, He X, Xu P, Huang S, Yuan Q, Zhang X, Wang L, Jiang K, Chen H, Li Z, Liu W, Wang S, Xu HE, Xu F
Nature (2023) 624 pp. 663-671 [ Pubmed: 37935377 DOI: doi:10.1038/s41586-023-06775-1 ]
- Trace amine-associated receptor 1 (39 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (37 kDa, Protein from Homo sapiens)
- Nanobody35 (15 kDa, Protein from Lama glama)
- Guanine nucleotide-binding protein g(s) subunit alpha isoforms short (28 kDa, Protein from Homo sapiens)
- Cryo-em structure of the ro5256390-taar1 complex (Complex from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
- (4s)-4-[(2s)-2-phenylbutyl]-1,3-oxazolidin-2-imine (218 Da, Ligand)
Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value
Resolution: 2.59 Å
EM Method: Single-particle
Fitted PDBs: 8iwx
Q-score: 0.466
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at high ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of protonated LHCII in detergent solution at low pH value
Resolution: 2.68 Å
EM Method: Single-particle
Fitted PDBs: 8iwy
Q-score: 0.572
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at low ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of unprotonated LHCII in detergent solution at low pH value
Resolution: 2.52 Å
EM Method: Single-particle
Fitted PDBs: 8iwz
Q-score: 0.563
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at low ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of unprotonated LHCII nanodisc at high pH value
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 8ix0
Q-score: 0.541
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at high ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of protonated LHCII nanodisc at low pH value
Resolution: 2.63 Å
EM Method: Single-particle
Fitted PDBs: 8ix1
Q-score: 0.526
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at low ph value (120 kDa, Complex from Spinacia)
Cryo-EM structure of unprotonated LHCII nanodisc at low pH value
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8ix2
Q-score: 0.531
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at low ph value (120 kDa, Complex from Spinacia)
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8e57
Q-score: 0.534
Yao X, Gao S, Wang J, Li Z, Huang J, Wang Y, Wang Z, Chen J, Fan X, Wang W, Jin X, Pan X, Yu Y, Lagrutta A, Yan N
Cell (2022) 185 pp. 4801-4810.e13 [ Pubmed: 36417914 DOI: doi:10.1016/j.cell.2022.10.024 ]
- (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone (645 Da, Ligand)
- Propan-2-yl (2s)-2-{[(s)-{[(2r,3s,4r,5r)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2h)-yl)-4-ethynyl-3-hydroxy-4-methyloxolan-2-yl]methoxy}(phenoxy)phosphoryl]amino}propanoate (non-preferred name) (535 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Voltage-dependent calcium channel gamma-1 subunit (25 kDa, Protein from Oryctolagus cuniculus)
- Voltage-dependent calcium channel subunit alpha-2/delta-1 (125 kDa, Protein from Oryctolagus cuniculus)
- Cav1.1 (Complex from Oryctolagus cuniculus)
- Voltage-dependent l-type calcium channel subunit alpha-1s (212 kDa, Protein from Oryctolagus cuniculus)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8e58
Q-score: 0.533
Yao X, Gao S, Wang J, Li Z, Huang J, Wang Y, Wang Z, Chen J, Fan X, Wang W, Jin X, Pan X, Yu Y, Lagrutta A, Yan N
Cell (2022) 185 pp. 4801-4810.e13 [ Pubmed: 36417914 DOI: doi:10.1016/j.cell.2022.10.024 ]
- (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone (645 Da, Ligand)
- Propan-2-yl (2s)-2-{[(s)-{[(2r,3s,4r,5r)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2h)-yl)-4-ethynyl-3-hydroxy-4-methyloxolan-2-yl]methoxy}(phenoxy)phosphoryl]amino}propanoate (non-preferred name) (535 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Voltage-dependent calcium channel gamma-1 subunit (25 kDa, Protein from Oryctolagus cuniculus)
- Voltage-dependent calcium channel subunit alpha-2/delta-1 (125 kDa, Protein from Oryctolagus cuniculus)
- Cav1.1 (Complex from Oryctolagus cuniculus)
- Voltage-dependent l-type calcium channel subunit alpha-1s (212 kDa, Protein from Oryctolagus cuniculus)
Cryo-EM structure of mouse temsirolimus/PI(3,5)P2-bound TRPML1 channel at 2.11 Angstrom resolution
Resolution: 2.11 Å
EM Method: Single-particle
Fitted PDBs: 7sq9
Q-score: 0.69
Gan N, Han Y, Zeng W, Wang Y, Xue J, Jiang Y
PNAS (2022) 119 [ DOI: doi:10.1073/pnas.2120404119 Pubmed: 35131932 ]
- (2r)-3-{[(s)-hydroxy{[(1s,2r,3r,4s,5s,6r)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate (746 Da, Ligand)
- Mucolipin-1 (65 kDa, Protein from Mus musculus)
- (1r,2r,4s)-4-{(2r)-2-[(3s,6r,7e,9r,10r,12r,14s,15e,17e,19e,21s,23s,26r,27r,30s,34as)-9,27-dihydroxy-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-1,5,11,28,29-pentaoxo-1,4,5,6,9,10,11,12,13,14,21,22,23,24,25,26,27,28,29,31,32,33,34,34a-tetracosahydro-3h-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontin-3-yl]propyl}-2-methoxycyclohexyl 3-hydroxy-2-(hydroxymethyl)-2-methylpropanoate (1 kDa, Ligand)
- Sodium ion (22 Da, Ligand)
- Mouse transient receptor potential-mucolipin 1 apo closed state (Complex from Mus musculus)
- Water (18 Da, Ligand)