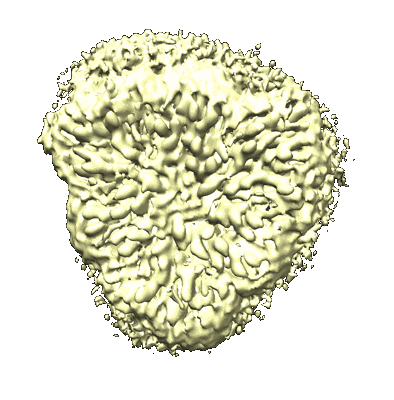
Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value
Resolution: 2.59 Å
EM Method: Single-particle
Fitted PDBs: 8iwx
Q-score: 0.466
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at high ph value (120 kDa, Complex from Spinacia oleracea)
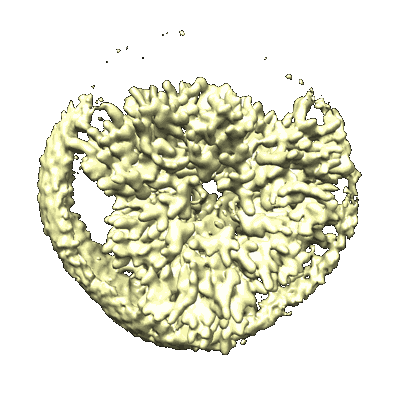
Cryo-EM structure of protonated LHCII in detergent solution at low pH value
Resolution: 2.68 Å
EM Method: Single-particle
Fitted PDBs: 8iwy
Q-score: 0.572
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at low ph value (120 kDa, Complex from Spinacia oleracea)
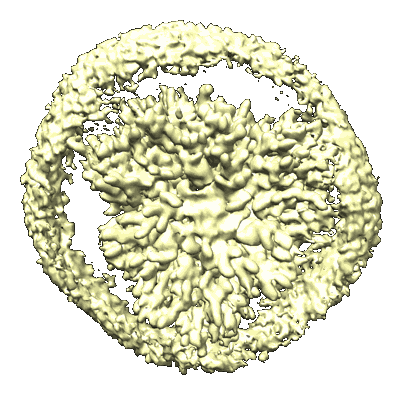
Cryo-EM structure of unprotonated LHCII nanodisc at high pH value
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 8ix0
Q-score: 0.541
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at high ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of protonated LHCII nanodisc at low pH value
Resolution: 2.63 Å
EM Method: Single-particle
Fitted PDBs: 8ix1
Q-score: 0.526
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at low ph value (120 kDa, Complex from Spinacia)
Cryo-EM structure of unprotonated LHCII nanodisc at low pH value
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8ix2
Q-score: 0.531
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at low ph value (120 kDa, Complex from Spinacia)
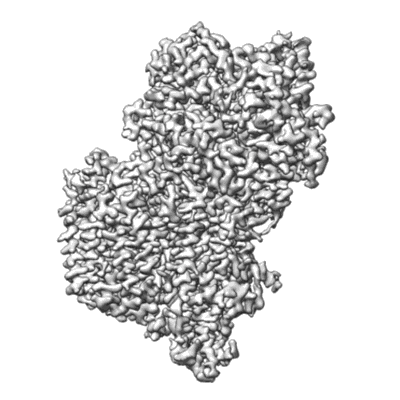
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 7bvc
Q-score: 0.564
Zhang L, Zhao Y, Gao Y, Wu L, Gao R, Zhang Q, Wang Y, Wu C, Wu F, Gurcha SS, Veerapen N, Batt SM, Zhao W, Qin L, Yang X, Wang M, Zhu Y, Zhang B, Bi L, Zhang X, Yang H, Guddat LW, Xu W, Wang Q, Li J, Besra GS, Rao Z
Science (2020) 368 pp. 1211-1219 [ DOI: doi:10.1126/science.aba9102 Pubmed: 32327601 ]
- Mycobacterium smegmatis arabinosyltransferase emba-embb-acpm2 in complex with ethambutol (Complex)
- Phosphate ion (94 Da, Ligand)
- Ethambutol (204 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Integral membrane indolylacetylinositol arabinosyltransferase embb (119 kDa, Protein from Mycolicibacterium smegmatis MC2 155)
- Cardiolipin (1 kDa, Ligand)
- [(2z,6e,10e,14z,18e,22z,26z)-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,10,14,18,22,26,30,34,38-decaenyl] [(2s,3s,4s,5r)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl] hydrogen phosphate (911 Da, Ligand)
- Acpm (Complex from Mycolicibacterium smegmatis MC2 155)
- 4'-phosphopantetheine (358 Da, Ligand)
- Integral membrane indolylacetylinositol arabinosyltransferase emba (116 kDa, Protein from Mycolicibacterium smegmatis MC2 155)
- Emba-embb (Complex from Mycolicibacterium smegmatis MC2 155)
- Meromycolate extension acyl carrier protein (10 kDa, Protein from Mycolicibacterium smegmatis MC2 155)
Cryo-EM density map of Alternative Complex III from Flavobacterium johnsoniae (Wild Type)
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6btm
Q-score: 0.482
Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosler J, Tajkhorshid E, Rubinstein JL, Gennis RB
Nature (2018) 557 pp. 123-126 [ DOI: doi:10.1038/s41586-018-0061-y Pubmed: 29695868 ]
- Fe3-s4 cluster (295 Da, Ligand)
- Alternative complex iii subunit f (52 kDa, Protein from Flavobacterium johnsoniae UW101)
- Decanoic acid (172 Da, Ligand)
- Alternative complex iii subunit e (18 kDa, Protein from Flavobacterium johnsoniae UW101)
- (2s)-3-hydroxypropane-1,2-diyl ditetradecanoate (512 Da, Ligand)
- Alternative complex iii subunit d (19 kDa, Protein from Flavobacterium johnsoniae UW101)
- Alternative complex iii subunit a (48 kDa, Protein from Flavobacterium johnsoniae UW101)
- Iron/sulfur cluster (351 Da, Ligand)
- Aciii-cyt aa3 supercomplex (Complex from Flavobacterium johnsoniae UW101)
- Heme c (618 Da, Ligand)
- Alternative complex iii subunit c (53 kDa, Protein from Flavobacterium johnsoniae UW101)
- Alternative complex iii subunit b (102 kDa, Protein from Flavobacterium johnsoniae UW101)