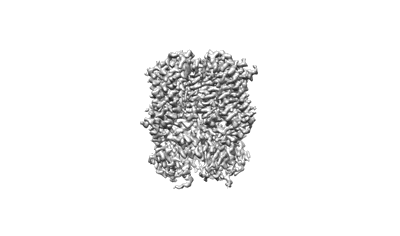
Cryo-EM structure of human HCN3 channel in apo state
Resolution: 2.72 Å
EM Method: Single-particle
Fitted PDBs: 8inz
Q-score: 0.526
Yu B, Lu Q, Li J, Cheng X, Hu H, Li Y, Che T, Hua Y, Jiang H, Zhang Y, Xian C, Yang T, Fu Y, Chen Y, Nan W, McCormick PJ, Xiong B, Duan J, Zeng B, Li Y, Fu Y, Zhang J
J Biol Chem (2024) 300 pp. 107288-107288 [ Pubmed: 38636662 DOI: doi:10.1016/j.jbc.2024.107288 ]
- 4-[[(2~{s},4~{a}~{r},6~{s},8~{a}~{s})-6-[(4~{s},5~{r})-4-[(2~{s})-butan-2-yl]-5,9-dimethyl-decyl]-4~{a}-methyl-2,3,4,5,6,7,8,8~{a}-octahydro-1~{h}-naphthalen-2-yl]oxy]-4-oxidanylidene-butanoic acid (492 Da, Ligand)
- Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (86 kDa, Protein from Homo sapiens)
- Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Complex from Homo sapiens)
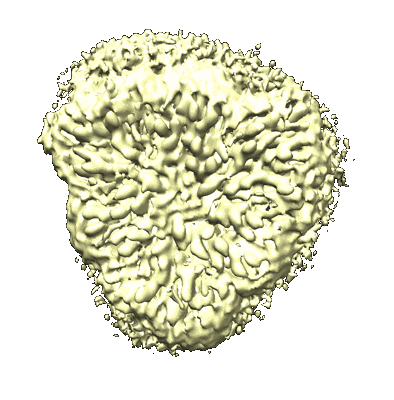
Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value
Resolution: 2.59 Å
EM Method: Single-particle
Fitted PDBs: 8iwx
Q-score: 0.466
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at high ph value (120 kDa, Complex from Spinacia oleracea)
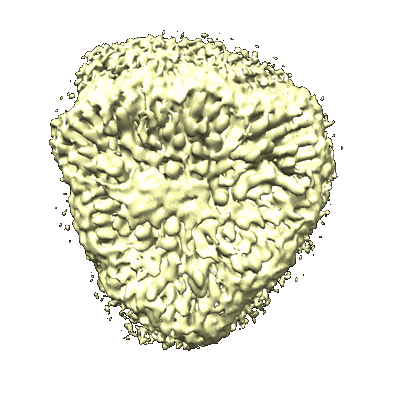
Cryo-EM structure of protonated LHCII in detergent solution at low pH value
Resolution: 2.68 Å
EM Method: Single-particle
Fitted PDBs: 8iwy
Q-score: 0.572
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at low ph value (120 kDa, Complex from Spinacia oleracea)
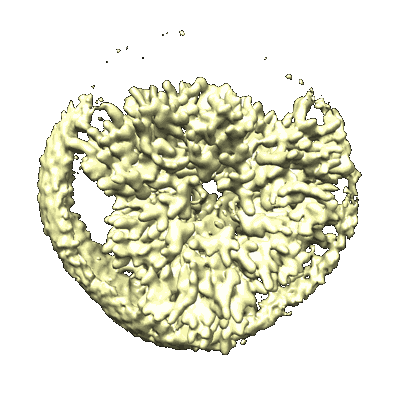
Cryo-EM structure of unprotonated LHCII in detergent solution at low pH value
Resolution: 2.52 Å
EM Method: Single-particle
Fitted PDBs: 8iwz
Q-score: 0.563
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at low ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of unprotonated LHCII nanodisc at high pH value
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 8ix0
Q-score: 0.541
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at high ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of protonated LHCII nanodisc at low pH value
Resolution: 2.63 Å
EM Method: Single-particle
Fitted PDBs: 8ix1
Q-score: 0.526
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at low ph value (120 kDa, Complex from Spinacia)
Cryo-EM structure of unprotonated LHCII nanodisc at low pH value
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8ix2
Q-score: 0.531
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at low ph value (120 kDa, Complex from Spinacia)
Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein
Resolution: 2.85 Å
EM Method: Single-particle
Fitted PDBs: 8imx
Q-score: 0.567
Xu Y, Li T, Zhou Z, Hong J, Chao Y, Zhu Z, Zhang Y, Qu Q, Li D
Nat Commun (2023) 14 pp. 5520-5520 [ DOI: doi:10.1038/s41467-023-41281-y Pubmed: 37684232 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Digitonin (1 kDa, Ligand)
- Gpi-anchor transamidase,gfp-like fluorescent chromoprotein cfp484 (73 kDa, Protein from Clavularia sp.)
- [(2r)-1-[[(1s,2r,3r,4s,5s,6r)-2-hexadecanoyloxy-3,4,5,6-tetrakis(oxidanyl)cyclohexyl]oxy-oxidanyl-phosphoryl]oxy-3-octadecoxy-propan-2-yl] (5z,8z,11z,14z)-icosa-5,8,11,14-tetraenoate (1 kDa, Ligand)
- Cholesterol hemisuccinate (486 Da, Ligand)
- Ul16-binding protein 2,gfp-like fluorescent chromoprotein cfp484 (35 kDa, Protein from Homo sapiens)
- Phosphatidylinositol glycan anchor biosynthesis class u protein,gfp-like fluorescent chromoprotein cfp484 (77 kDa, Protein from Clavularia sp.)
- 2-amino-2-deoxy-alpha-d-glucopyranose (179 Da, Ligand)
- Gpi transamidase component pig-s,gfp-like fluorescent chromoprotein cfp484 (90 kDa, Protein from Clavularia sp.)
- Gpi transamidase component pig-t,gfp-like fluorescent chromoprotein cfp484 (94 kDa, Protein from Clavularia sp.)
- [(2r)-1-hexadecanoyloxy-3-[[3-[[(2r)-3-hexadecanoyloxy-2-[(z)-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-2-oxidanyl-propoxy]-oxidanyl-phosphoryl]oxy-propan-2-yl] (z)-octadec-9-enoate (1 kDa, Ligand)
- Complex of gpi-t with a chimeric gpi-anchored protein (469 MDa, Complex from Homo sapiens)
- Glycosylphosphatidylinositol anchor attachment 1 protein,gfp-like fluorescent chromoprotein cfp484 (96 kDa, Protein from Clavularia sp.)
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (760 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- [(2~{r})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{z})-octadec-9-enoate (717 Da, Ligand)
- 2-azanylethyl [(2r,3s,4s,5s,6s)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate (303 Da, Ligand)
- Met-ser-ser peptide (323 Da, Protein from synthetic construct)
- Calcium ion (40 Da, Ligand)
- 2-azanylethyl [(2~{s},3~{s},4~{s},5~{s},6~{r})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate (303 Da, Ligand)
cryo-EM structure of the human respiratory complex II
Resolution: 2.86 Å
EM Method: Single-particle
Fitted PDBs: 8gs8
Q-score: 0.577
Du Z, Zhou X, Lai Y, Xu J, Zhang Y, Zhou S, Feng Z, Yu L, Tang Y, Wang W, Yu L, Tian C, Ran T, Chen H, Guddat LW, Liu F, Gao Y, Rao Z, Gong H
PNAS (2023) 120 pp. e2216713120-e2216713120 [ DOI: doi:10.1073/pnas.2216713120 Pubmed: 37098072 ]
- Fe3-s4 cluster (295 Da, Ligand)
- Ubiquinone-1 (250 Da, Ligand)
- Succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial (17 kDa, Protein from Homo sapiens)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- (1s)-2-{[(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(palmitoyloxy)methyl]ethyl stearate (720 Da, Ligand)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- The human mitochondrial complex ii, composed of sdha, sdhb, sdhc and sdhd (Complex from Homo sapiens)
- Succinate dehydrogenase cytochrome b560 subunit, mitochondrial (18 kDa, Protein from Homo sapiens)
- Iron/sulfur cluster (351 Da, Ligand)
- Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial (31 kDa, Protein from Homo sapiens)
- Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial (72 kDa, Protein from Homo sapiens)
- Flavin-adenine dinucleotide (785 Da, Ligand)
human KCNQ1-CaM-ML277-PIP2 complex in state B
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 7xnn
Q-score: 0.428
Ma D, Zhong L, Yan Z, Yao J, Zhang Y, Ye F, Huang Y, Lai D, Yang W, Hou P, Guo J
PNAS (2022) 119 pp. e2207067119-e2207067119 [ DOI: doi:10.1073/pnas.2207067119 Pubmed: 36763058 ]
- Potassium voltage-gated channel subfamily kqt member 1 (76 kDa, Protein from Homo sapiens)
- [(2r)-2-octanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate (746 Da, Ligand)
- Calmodulin-3 (19 kDa, Protein from Homo sapiens)
- Potassium ion (39 Da, Ligand)
- (2r)-n-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide (471 Da, Ligand)
- Human kcnq1-cam-ml277-pip2 complex in state b (Cellular component from Homo sapiens)