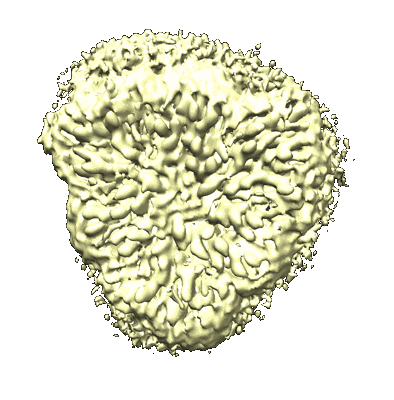
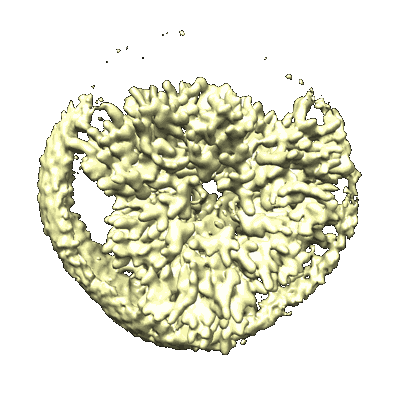
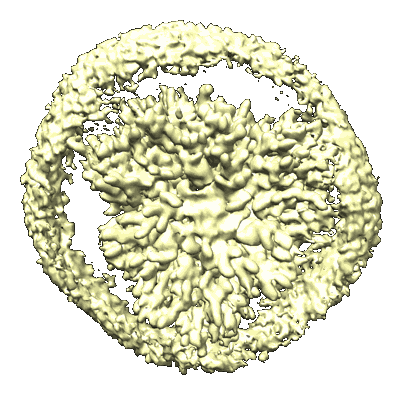
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with GABA and puerarin
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 9eqg
Q-score: 0.627
Lyu Q, Xue W, Liu R, Ma Q, Kasaragod VB, Sun S, Li Q, Chen Y, Yuan M, Yang Y, Zhang B, Nie A, Jia S, Shen C, Gao P, Rong W, Yu C, Bi Y, Zhang C, Nan F, Ning G, Rao Z, Yang X, Wang J, Wang W
Nature (2024) 634 pp. 936-943 [ Pubmed: 39261733 DOI: doi:10.1038/s41586-024-07929-5 ]
- Gamma-aminobutyric acid receptor subunit beta-3 (54 kDa, Protein from Homo sapiens)
- Hexane (86 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 1,2-dilauroyl-sn-glycero-3-phosphate (535 Da, Ligand)
- Water (18 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Gamma-amino-butanoic acid (103 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit gamma-2 (56 kDa, Protein from Homo sapiens)
- [(2r)-1-octadecanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8z)-icosa-5,8,11,14-tetraenoate (1 kDa, Ligand)
- Hexadecane (226 Da, Ligand)
- Decane (142 Da, Ligand)
- (1r)-2-{[(s)-{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9z)-octadec-9-enoate (749 Da, Ligand)
- Puerarin (416 Da, Ligand)
- Cryoem structure of human full-length alpha1beta3gamma2l gaba(a)r in complex with gaba and puerarin (250 kDa, Complex from Homo sapiens)
- Chloride ion (35 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit alpha-1 (52 kDa, Protein from Homo sapiens)
Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value
Resolution: 2.59 Å
EM Method: Single-particle
Fitted PDBs: 8iwx
Q-score: 0.466
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at high ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of protonated LHCII in detergent solution at low pH value
Resolution: 2.68 Å
EM Method: Single-particle
Fitted PDBs: 8iwy
Q-score: 0.572
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at low ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of unprotonated LHCII nanodisc at high pH value
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 8ix0
Q-score: 0.541
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at high ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of protonated LHCII nanodisc at low pH value
Resolution: 2.63 Å
EM Method: Single-particle
Fitted PDBs: 8ix1
Q-score: 0.526
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at low ph value (120 kDa, Complex from Spinacia)
Cryo-EM structure of unprotonated LHCII nanodisc at low pH value
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8ix2
Q-score: 0.531
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at low ph value (120 kDa, Complex from Spinacia)
Structure of Mycobacterium smegmatis succinate dehydrogenase 2
Resolution: 2.84 Å
EM Method: Single-particle
Fitted PDBs: 6lum
Q-score: 0.561
Gong H, Gao Y, Zhou X, Xiao Y, Wang W, Tang Y, Zhou S, Zhang Y, Ji W, Yu L, Tian C, Lam SM, Shui G, Guddat LW, Wong LL, Wang Q, Rao Z
Nat Commun (2020) 11 pp. 4245-4245 [ DOI: doi:10.1038/s41467-020-18011-9 Pubmed: 32843629 ]
- Mycobaterium smegmatis succinate dehydrogenase 2 (400 kDa, Complex from Mycolicibacterium smegmatis MC2 51)
- Succinate dehydrogenase subunit a (64 kDa, Protein from Mycolicibacterium smegmatis MC2 51)
- Menaquinone-9 (785 Da, Ligand)
- Fe3-s4 cluster (295 Da, Ligand)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- (1s)-2-{[(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(palmitoyloxy)methyl]ethyl stearate (720 Da, Ligand)
- Succinate dehydrogenase subunit f (3 kDa, Protein from Mycolicibacterium smegmatis MC2 51)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- 1,2-diacyl-sn-glycero-3-phosphoinositol (836 Da, Ligand)
- 2-(hexadecanoyloxy)-1-[(phosphonooxy)methyl]ethyl hexadecanoate (648 Da, Ligand)
- Succinate dehydrogenase subunit d (19 kDa, Protein from Mycolicibacterium smegmatis MC2 51)
- Iron/sulfur cluster (351 Da, Ligand)
- Succinate dehydrogenase subunit c (15 kDa, Protein from Mycolicibacterium smegmatis MC2 51)
- Cardiolipin (1 kDa, Ligand)
- Succinate dehydrogenase subunit b (29 kDa, Protein from Mycolicibacterium smegmatis MC2 51)
- Flavin-adenine dinucleotide (785 Da, Ligand)
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7bwr
Q-score: 0.419
Zhang L, Zhao Y, Gao R, Li J, Yang X, Gao Y, Zhao W, Gurcha SS, Veerapen N, Batt SM, Besra KK, Xu W, Bi L, Zhang X, Guddat LW, Yang H, Wang Q, Besra GS, Rao Z
Protein Cell (2020) 11 pp. 505-517 [ DOI: doi:10.1007/s13238-020-00726-6 Pubmed: 32363534 ]
- Calcium ion (40 Da, Ligand)
- Integral membrane indolylacetylinositol arabinosyltransferase embb (116 kDa, Protein from Mycolicibacterium smegmatis MC2 155)
- [(2z,6e,10e,14z,18e,22z,26z)-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,10,14,18,22,26,30,34,38-decaenyl] [(2s,3s,4s,5r)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl] hydrogen phosphate (911 Da, Ligand)
- Mycobacterial arabinosyltransferase complex embb2-acpm2 (254 MDa, Complex from Mycolicibacterium smegmatis MC2 155)
- Meromycolate extension acyl carrier protein (10 kDa, Protein from Mycolicibacterium smegmatis MC2 155)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 7bvc
Q-score: 0.564
Zhang L, Zhao Y, Gao Y, Wu L, Gao R, Zhang Q, Wang Y, Wu C, Wu F, Gurcha SS, Veerapen N, Batt SM, Zhao W, Qin L, Yang X, Wang M, Zhu Y, Zhang B, Bi L, Zhang X, Yang H, Guddat LW, Xu W, Wang Q, Li J, Besra GS, Rao Z
Science (2020) 368 pp. 1211-1219 [ DOI: doi:10.1126/science.aba9102 Pubmed: 32327601 ]
- Mycobacterium smegmatis arabinosyltransferase emba-embb-acpm2 in complex with ethambutol (Complex)
- Phosphate ion (94 Da, Ligand)
- Ethambutol (204 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Integral membrane indolylacetylinositol arabinosyltransferase embb (119 kDa, Protein from Mycolicibacterium smegmatis MC2 155)
- Cardiolipin (1 kDa, Ligand)
- [(2z,6e,10e,14z,18e,22z,26z)-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,10,14,18,22,26,30,34,38-decaenyl] [(2s,3s,4s,5r)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl] hydrogen phosphate (911 Da, Ligand)
- Acpm (Complex from Mycolicibacterium smegmatis MC2 155)
- 4'-phosphopantetheine (358 Da, Ligand)
- Integral membrane indolylacetylinositol arabinosyltransferase emba (116 kDa, Protein from Mycolicibacterium smegmatis MC2 155)
- Emba-embb (Complex from Mycolicibacterium smegmatis MC2 155)
- Meromycolate extension acyl carrier protein (10 kDa, Protein from Mycolicibacterium smegmatis MC2 155)