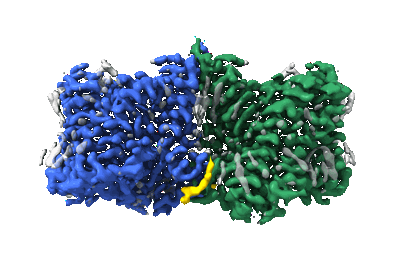
3D structure of Y-50 TCR-TMM-CD1b ternary complex
Resolution: 3.18 Å
EM Method: Single-particle
Fitted PDBs: 8zox
Q-score: 0.506
Sakai Y, Asa M, Hirose M, Kusuhara W, Fujiwara N, Tamashima H, Ikazaki T, Oka S, Kuraba K, Tanaka K, Yoshiyama T, Nagae M, Hoshino Y, Motooka D, Van Rhijn I, Lu X, Ishikawa E, Moody DB, Kato T, Inuki S, Hirai G, Yamasaki S
J Clin Inv (2024) [ DOI: doi:10.1172/JCI185443 Pubmed: 39718834 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Beta-2-microglobulin (11 kDa, Protein from Homo sapiens)
- Alpha-d-glucopyranose (180 Da, Ligand)
- T-cell surface glycoprotein cd1b (31 kDa, Protein from Homo sapiens)
- Y-50 tcr beta (28 kDa, Protein from Homo sapiens)
- [(2r,3s,4s,5r,6s)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl (2r,3r)-3-oxidanyl-2-tetradecyl-octadecanoate (658 Da, Ligand)
- Tetracosyl palmitate (593 Da, Ligand)
- Y-50 tcr alpha (23 kDa, Protein from Homo sapiens)
- Ternary complex of y-50tcr,tmm and cd1b (Complex from Homo sapiens)
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with GABA and puerarin
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 9eqg
Q-score: 0.627
Lyu Q, Xue W, Liu R, Ma Q, Kasaragod VB, Sun S, Li Q, Chen Y, Yuan M, Yang Y, Zhang B, Nie A, Jia S, Shen C, Gao P, Rong W, Yu C, Bi Y, Zhang C, Nan F, Ning G, Rao Z, Yang X, Wang J, Wang W
Nature (2024) 634 pp. 936-943 [ Pubmed: 39261733 DOI: doi:10.1038/s41586-024-07929-5 ]
- Gamma-aminobutyric acid receptor subunit beta-3 (54 kDa, Protein from Homo sapiens)
- Hexane (86 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 1,2-dilauroyl-sn-glycero-3-phosphate (535 Da, Ligand)
- Water (18 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Gamma-amino-butanoic acid (103 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit gamma-2 (56 kDa, Protein from Homo sapiens)
- [(2r)-1-octadecanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8z)-icosa-5,8,11,14-tetraenoate (1 kDa, Ligand)
- Hexadecane (226 Da, Ligand)
- Decane (142 Da, Ligand)
- (1r)-2-{[(s)-{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9z)-octadec-9-enoate (749 Da, Ligand)
- Puerarin (416 Da, Ligand)
- Cryoem structure of human full-length alpha1beta3gamma2l gaba(a)r in complex with gaba and puerarin (250 kDa, Complex from Homo sapiens)
- Chloride ion (35 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit alpha-1 (52 kDa, Protein from Homo sapiens)
Cryo-EM structure of the mouse TRPM8 channel in complex with PI(4,5)P2 and Ca2+
Yin Y, Park CG, Zhang F, G Fedor J, Feng S, Suo Y, Im W, Lee SY
Sci Adv (2024) 10 pp. eadp2211-eadp2211 [ Pubmed: 39093967 DOI: doi:10.1126/sciadv.adp2211 ]
- Transient receptor potential cation channel subfamily m member 8 (Complex from Mus musculus)
- [(2r)-2-octanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate (746 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Transient receptor potential cation channel subfamily m member 8 (131 kDa, Protein from Mus musculus)
Structure of AE2 in complex with PIP2
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8jni
Q-score: 0.532
Zhang W, Ding D, Lu Y, Chen H, Jiang P, Zuo P, Wang G, Luo J, Yin Y, Luo J, Yin Y
Nat Commun (2024) 15 pp. 759-759 [ Pubmed: 38272905 DOI: doi:10.1038/s41467-024-44966-0 ]
- Anion exchange protein 2, isoform a (Complex from Homo sapiens)
- [(2r)-1-octadecanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8z)-icosa-5,8,11,14-tetraenoate (1 kDa, Ligand)
- Anion exchange protein 2 (137 kDa, Protein from Homo sapiens)
- Chloride ion (35 Da, Ligand)
PROTAC-mediated complex of KRAS with VHL/Elongin-B/Elongin-C/Cullin-2/Rbx1
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8qu8
Q-score: 0.306
Popow J, Farnaby W, Gollner A, Kofink C, Fischer G, Wurm M, Zollman D, Wijaya A, Mischerikow N, Hasenoehrl C, Prokofeva P, Arnhof H, Arce-Solano S, Bell S, Boeck G, Diers E, Frost AB, Goodwin-Tindall J, Karolyi-Oezguer J, Khan S, Klawatsch T, Koegl M, Kousek R, Kratochvil B, Kropatsch K, Lauber AA, McLennan R, Olt S, Peter D, Petermann O, Roessler V, Stolt-Bergner P, Strack P, Strauss E, Trainor N, Vetma V, Whitworth C, Zhong S, Quant J, Weinstabl H, Kuster B, Ettmayer P, Ciulli A
Science (2024) 385 pp. 1338-1347 [ DOI: doi:10.1126/science.adm8684 Pubmed: 39298590 ]
- Zinc ion (65 Da, Ligand)
- Gtpase kras (19 kDa, Protein from Homo sapiens)
- (2s,4r)-1-[(2s)-2-[4-[4-[(3s)-4-[4-[5-[(4s)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5h-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-n-[(1r)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide (1 kDa, Ligand)
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Kras/acbi3/vhl/elob/eloc/cul2/rbx1 (168 kDa, Complex from Homo sapiens)
- Elongin-b (13 kDa, Protein from Homo sapiens)
- Elongin-c (12 kDa, Protein from Homo sapiens)
- Cullin-2 (86 kDa, Protein from Homo sapiens)
- E3 ubiquitin-protein ligase rbx1, n-terminally processed (12 kDa, Protein from Homo sapiens)
- Von hippel-lindau disease tumor suppressor (24 kDa, Protein from Homo sapiens)
Cryo-EM structure of human BTR1 in the outward-facing state.
Resolution: 2.94 Å
EM Method: Single-particle
Fitted PDBs: 7x1i
Q-score: 0.578
Lu Y, Zuo P, Chen H, Shan H, Wang W, Dai Z, Xu H, Chen Y, Liang L, Ding D, Jin Y, Yin Y
Nat Commun (2023) 14 pp. 6157-6157 [ Pubmed: 37788993 DOI: doi:10.1038/s41467-023-41924-0 ]
- Isoform 1 of solute carrier family 4 member 11 (99 kDa, Protein from Homo sapiens)
- [(2r)-1-octadecanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8z)-icosa-5,8,11,14-tetraenoate (1 kDa, Ligand)
- Solute carrier family 4 member 11, isoform b (199 kDa, Complex from Homo sapiens)
Cryo-EM structure of human BTR1 in the outward-facing state in the presence of NH4Cl.
Resolution: 2.84 Å
EM Method: Single-particle
Fitted PDBs: 7x1j
Q-score: 0.572
Lu Y, Zuo P, Chen H, Shan H, Wang W, Dai Z, Xu H, Chen Y, Liang L, Ding D, Jin Y, Yin Y
Nat Commun (2023) 14 pp. 6157-6157 [ Pubmed: 37788993 DOI: doi:10.1038/s41467-023-41924-0 ]
- Isoform 1 of solute carrier family 4 member 11 (99 kDa, Protein from Homo sapiens)
- [(2r)-1-octadecanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8z)-icosa-5,8,11,14-tetraenoate (1 kDa, Ligand)
- Solute carrier family 4 member 11, isoform b (199 kDa, Complex from Homo sapiens)
Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value
Resolution: 2.59 Å
EM Method: Single-particle
Fitted PDBs: 8iwx
Q-score: 0.466
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at high ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of protonated LHCII in detergent solution at low pH value
Resolution: 2.68 Å
EM Method: Single-particle
Fitted PDBs: 8iwy
Q-score: 0.572
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii in detergent solution at low ph value (120 kDa, Complex from Spinacia oleracea)
Cryo-EM structure of unprotonated LHCII nanodisc at high pH value
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 8ix0
Q-score: 0.541
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Lhcii nanodsic at high ph value (120 kDa, Complex from Spinacia oleracea)