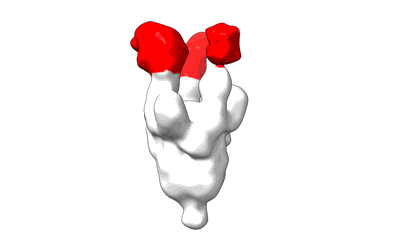
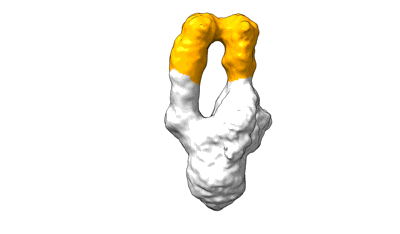
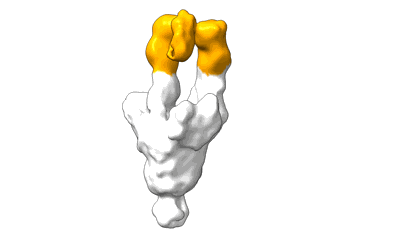
Locally refined region of SARS-CoV-2 spike in complex with antibodies 9G11 and 3E2.
Resolution: 3.46 Å
EM Method: Single-particle
Fitted PDBs: 9lae
Q-score: 0.522
Jiang W, Jiang Y, Sun H, Deng T, Yu K, Fang Q, Ge H, Lan M, Lin Y, Fang Z, Zhang Y, Zhou L, Li T, Yu H, Zheng Q, Li S, Xia N, Gu Y
Emerg Microbes Infect (2024) 13 pp. 2406300- [ DOI: doi:10.1038/s41467-023-39265-z Pubmed: 39470577 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike in complex with antibodies 9g11 and 3e2 (Complex)
- Light chain of 9g11 (11 kDa, Protein from Mus musculus)
- Heavy chain of 3e2 (13 kDa, Protein from Mus musculus)
- The fab of 3e2 (Complex from Mus musculus)
- Heavy chain of 9g11 (13 kDa, Protein from Mus musculus)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
- Light chain of 3e2 (11 kDa, Protein from Mus musculus)
- The fab of 9g11 (Complex from Mus musculus)
- Spike protein s1 (25 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 Spike in complex with antibodies 3E2, 9G11 and 13H7.
Structure of SARS-CoV-2 Spike in complex with antibodies 3E2, 9G11 and 13H7 (C1)
Jiang W, Jiang Y, Sun H, Deng T, Yu K, Fang Q, Ge H, Lan M, Lin Y, Fang Z, Zhang Y, Zhou L, Li T, Yu H, Zheng Q, Li S, Xia N, Gu Y
Emerg Microbes Infect (2024) 13 pp. 2406300-2406300 [ DOI: doi:10.1080/22221751.2024.2406300 Pubmed: 39470577 ]
- The fab of 13h7 (Complex from Mus)
- Sars-cov-2 spike (Complex from Homo sapiens)
- The fab of 9g11 (Complex from Mus)
- The fab of 3e2 (Complex)
- Sars-cov-2 spike in complex with 3e2, 9g11 and 13h7 fab (Complex)
Cryo-EM structure of SARS-CoV-2 spike in complex with VHH14
Resolution: 4.6 Å
EM Method: Single-particle
Fitted PDBs: 7xy3
Q-score: 0.278
Wang K, Cao D, Liu L, Fan X, Wang H, Zhang X, Yang P
To Be Published
- Vhh14 (17 kDa, Protein from Camelus bactrianus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (125 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Vhh14 (Complex from Camelus bactrianus)
- Sars-cov-2 spike in complex with vhh14 (Complex)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of SARS-CoV-2 spike in complex with VHH21
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7xy4
Q-score: 0.43
Wang K, Cao D, Liu L, Fan X, Wang H, Zhang X, Yang P
To Be Published
- Vhh21 (Complex from Camelus bactrianus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (125 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Vhh21 (16 kDa, Protein from Camelus bactrianus)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike in complex with vhh21 (Complex)
Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7yc5
Q-score: 0.284
Kim JW, Heo K, Kim HJ, Yoo Y, Cho HS, Jang HJ, Lee HY, Ko IY, Woo JR, Cho YB, Lee JH, Yang HR, Shin HG, Choi HL, Hwang K, Kim S, Kim H, Chun K, Lee S
Antiviral Res (2023) 212 pp. 105576-105576 [ Pubmed: 36870394 DOI: doi:10.1016/j.antiviral.2023.105576 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (133 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Light chain from k202.b, bispecific antibody (49 kDa, Protein from Homo sapiens)
- Heavy chain from k202.b, bispecific antibody (49 kDa, Protein from Homo sapiens)
- Cryo-em structure of sars-cov-2 spike in complex with k202.b bispecific antibody (Complex from Severe acute respiratory syndrome coronavirus 2)
- K202.b bispecific antibody (Complex)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-093
Callaway HM, Hastie KM, Schendel SL, Li H, Yu X, Shek J, Buck T, Hui S, Bedinger D, Troup C, Dennison SM, Li K, Alpert MD, Bailey CC, Benzeno S, Bonnevier JL, Chen JQ, Chen C, Cho H, Crompton PD, Dussupt V, Entzminger KC, Ezzyat Y, Fleming JK, Geukens N, Gilbert AE, Guan Y, Han X, Harvey CJ, Hatler JM, Howie B, Hu C, Huang A, Imbrechts M, Jin A, Kamachi N, Keitany G, Klinger M, Kolls JK, Krebs SJ, Li T, Luo F, Maruyama T, Meehl MA, Mendez-Rivera L, Musa A, Okumura CJ, Rubin BER, Sato AK, Shen M, Singh A, Song S, Tan J, Trimarchi JM, Upadhyay DP, Wang Y, Yu L, Yuan TZ, Yusko E, Peters B, Tomaras G, Saphire EO
Cell Rep (2023) 42 pp. 112014-112014 [ Pubmed: 36681898 DOI: doi:10.1016/j.celrep.2023.112014 ]
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-156
Callaway HM, Hastie KM, Schendel SL, Li H, Yu X, Shek J, Buck T, Hui S, Bedinger D, Troup C, Dennison SM, Li K, Alpert MD, Bailey CC, Benzeno S, Bonnevier JL, Chen JQ, Chen C, Cho H, Crompton PD, Dussupt V, Entzminger KC, Ezzyat Y, Fleming JK, Geukens N, Gilbert AE, Guan Y, Han X, Harvey CJ, Hatler JM, Howie B, Hu C, Huang A, Imbrechts M, Jin A, Kamachi N, Keitany G, Klinger M, Kolls JK, Krebs SJ, Li T, Luo F, Maruyama T, Meehl MA, Mendez-Rivera L, Musa A, Okumura CJ, Rubin BER, Sato AK, Shen M, Singh A, Song S, Tan J, Trimarchi JM, Upadhyay DP, Wang Y, Yu L, Yuan TZ, Yusko E, Peters B, Tomaras G, Saphire EO
Cell Rep (2023) 42 pp. 112014-112014 [ Pubmed: 36681898 DOI: doi:10.1016/j.celrep.2023.112014 ]
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-279
Callaway HM, Hastie KM, Schendel SL, Li H, Yu X, Shek J, Buck T, Hui S, Bedinger D, Troup C, Dennison SM, Li K, Alpert MD, Bailey CC, Benzeno S, Bonnevier JL, Chen JQ, Chen C, Cho H, Crompton PD, Dussupt V, Entzminger KC, Ezzyat Y, Fleming JK, Geukens N, Gilbert AE, Guan Y, Han X, Harvey CJ, Hatler JM, Howie B, Hu C, Huang A, Imbrechts M, Jin A, Kamachi N, Keitany G, Klinger M, Kolls JK, Krebs SJ, Li T, Luo F, Maruyama T, Meehl MA, Mendez-Rivera L, Musa A, Okumura CJ, Rubin BER, Sato AK, Shen M, Singh A, Song S, Tan J, Trimarchi JM, Upadhyay DP, Wang Y, Yu L, Yuan TZ, Yusko E, Peters B, Tomaras G, Saphire EO
Cell Rep (2023) 42 pp. 112014-112014 [ Pubmed: 36681898 DOI: doi:10.1016/j.celrep.2023.112014 ]
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-290
Callaway HM, Hastie KM, Schendel SL, Li H, Yu X, Shek J, Buck T, Hui S, Bedinger D, Troup C, Dennison SM, Li K, Alpert MD, Bailey CC, Benzeno S, Bonnevier JL, Chen JQ, Chen C, Cho H, Crompton PD, Dussupt V, Entzminger KC, Ezzyat Y, Fleming JK, Geukens N, Gilbert AE, Guan Y, Han X, Harvey CJ, Hatler JM, Howie B, Hu C, Huang A, Imbrechts M, Jin A, Kamachi N, Keitany G, Klinger M, Kolls JK, Krebs SJ, Li T, Luo F, Maruyama T, Meehl MA, Mendez-Rivera L, Musa A, Okumura CJ, Rubin BER, Sato AK, Shen M, Singh A, Song S, Tan J, Trimarchi JM, Upadhyay DP, Wang Y, Yu L, Yuan TZ, Yusko E, Peters B, Tomaras G, Saphire EO
Cell Rep (2023) 42 pp. 112014-112014 [ Pubmed: 36681898 DOI: doi:10.1016/j.celrep.2023.112014 ]