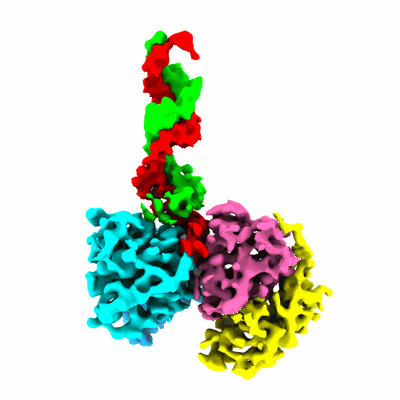
HIV-1 Vif in complex with human APOBEC3H, CBF-beta, ELOB, ELOC, CUL5, and RBX2
Ito F, Alvarez-Cabrera AL, Kim K, Zhou ZH, Chen XS
Nat Commun (2023) 14 pp. 5241-5241 [ DOI: doi:10.1038/s41467-023-40955-x Pubmed: 37640699 ]
- Elongin-c (Protein from Homo sapiens)
- Cullin-5 (Protein from Homo sapiens)
- Dna dc->du-editing enzyme apobec-3h (Protein from Homo sapiens)
- Virion infectivity factor (Protein from Human immunodeficiency virus 1)
- Ring-box protein 2 (Protein from Homo sapiens)
- Core-binding factor subunit beta (Protein from Homo sapiens)
- Hetero-heptameric complex of hiv-1 vif and human apobec3h, cbf-beta, elob, eloc, cul5, and rbx2 (Complex from Homo sapiens)
- Elongin-b (Protein from Homo sapiens)
Apo Hantaan virus polymerase core
Resolution: 3.27 Å
EM Method: Single-particle
Fitted PDBs: 8c4s
Q-score: 0.508
Durieux Trouilleton Q, Barata-Garcia S, Arragain B, Reguera J, Malet H
Nat Commun (2023) 14 pp. 2954-2954 [ Pubmed: 37221161 DOI: doi:10.1038/s41467-023-38555-w ]
- Apo hantaan virus polymerase core (246 kDa, Complex from Hantaan virus 76-118)
- Rna-directed rna polymerase l (249 kDa, Protein from Hantaan virus 76-118)
Resolution: 3.35 Å
EM Method: Single-particle
Fitted PDBs: 8oiu
Q-score: 0.465
Skalidis I, Kyrilis FL, Tuting C, Hamdi F, Trager TK, Belapure J, Hause G, Fratini M, O'Reilly FJ, Heilmann I, Rappsilber J, Kastritis PL
Commun Biol (2023) 6 pp. 552-552 [ Pubmed: 37217784 DOI: doi:10.1038/s42003-023-04885-0 ]
Full-length APOBEC3G in complex with HIV-1 Vif, CBF-beta, and fork RNA
Resolution: 3.57 Å
EM Method: Single-particle
Fitted PDBs: 8e40
Q-score: 0.343
Ito F, Alvarez-Cabrera AL, Liu S, Yang H, Shiriaeva A, Zhou ZH, Chen XS
Sci Adv (2023) 9 pp. eade3168-eade3168 [ Pubmed: 36598981 DOI: doi:10.1126/sciadv.ade3168 ]
- Virion infectivity factor (21 kDa, Protein from Human immunodeficiency virus 1)
- Rna (8 kDa, RNA from Escherichia coli)
- Dna dc->du-editing enzyme apobec-3g (45 kDa, Protein from Macaca mulatta)
- Zinc ion (65 Da, Ligand)
- Complex of apobec3g with hiv-1 vif and cbf-beta bound to fork rna (99 kDa, Complex from Human immunodeficiency virus 1)
- Rna (6 kDa, RNA from Escherichia coli)
- Core-binding factor subunit beta (18 kDa, Protein from Homo sapiens)
APOBEC3G in complex with HIV-1 Vif/CBF-beta/EloB/EloC/Cul5/Rbx2
Ito F, Alvarez-Cabrera AL, Liu S, Yang H, Shiriaeva A, Zhou ZH, Chen XS
Sci Adv (2023) 9 pp. eade3168-eade3168 [ Pubmed: 36598981 DOI: doi:10.1126/sciadv.ade3168 ]
- Elongin-c (Protein from Homo sapiens)
- Apobec3g (Protein from Macaca mulatta)
- Cullin-5 (Protein from Homo sapiens)
- Complex of apobec3g with hiv-1 vif, cbf-beta, elongin-b, elongin-c, cullin-5, and ring-box protein 2 (210 kDa, Complex from Human immunodeficiency virus 1)
- Virion infectivity factor (Protein from Human immunodeficiency virus 1)
- Ring-box protein 2 (Protein from Homo sapiens)
- Core-binding factor subunit beta (Protein from Homo sapiens)
- Elongin-b (Protein from Homo sapiens)
Immature 60S Ribosomal Subunit from C. thermophilum
Skalidis I, Kyrilis FL, Tuting C, Hamdi F, Chojnowski G, Kastritis PL
Structure (2022) 30 pp. 575-589.e6 [ Pubmed: 35093201 DOI: doi:10.1016/j.str.2022.01.001 ]
- Ul6 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Ul22 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Ul29 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Ul5 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Ul18 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- El20 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Ul13 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Ul24 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- El8 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Ul30 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- El21 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Native 60-mer core of pyruvate dehydrogenase complex (3 MDa, Complex from Chaetomium thermophilum var. thermophilum DSM 1495)
- El14 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- El15 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- El32 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- El6 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- El13 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- El18 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- El33 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- El36 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Ul4 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Ul15 (Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
Protein community member oxoglutarate dehydrogenase complex E2 core from C. thermophilum
Resolution: 4.38 Å
EM Method: Single-particle
Fitted PDBs: 7q5q
Q-score: 0.308
Skalidis I, Kyrilis FL, Tuting C, Hamdi F, Chojnowski G, Kastritis PL
Structure (2022) 30 pp. 575- [ Pubmed: 35093201 DOI: doi:10.1016/j.str.2022.01.001 ]
Protein community member pyruvate dehydrogenase complex E2 core from C. thermophilum
Resolution: 3.84 Å
EM Method: Single-particle
Fitted PDBs: 7q5r
Q-score: 0.21
Skalidis I, Kyrilis FL, Tuting C, Hamdi F, Chojnowski G, Kastritis PL
Structure (2022) 30 pp. 575- [ Pubmed: 35093201 DOI: doi:10.1016/j.str.2022.01.001 ]
Metabolon-embedded pyruvate dehydrogenase complex E2 core at near-atomic resolution
Resolution: 3.84 Å
EM Method: Single-particle
Fitted PDBs: 7ott
Q-score: 0.42
Tuting C, Kyrilis FL, Muller J, Sorokina M, Skalidis I, Hamdi F, Sadian Y, Kastritis PL
Nat Commun (2021) 12 pp. 6933-6933 [ DOI: doi:10.1038/s41467-021-27287-4 Pubmed: 34836937 ]
- Metabolon-embedded pyruvate dehydrogenase complex e2 core at near-atomic resolution (Complex from Chaetomium thermophilum var. thermophilum DSM 1495)
- Acetyltransferase component of pyruvate dehydrogenase complex (48 kDa, Protein from Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719))
Resolution: 6.9 Å
EM Method: Single-particle
Fitted PDBs: 7bgj
Q-score: 0.167
Kyrilis FL, Semchonok DA, Skalidis I, Tuting C, Hamdi F, O'Reilly FJ, Rappsilber J, Kastritis PL
Cell Rep (2021) 34 pp. 108727-108727 [ DOI: doi:10.1016/j.celrep.2021.108727 Pubmed: 33567276 ]