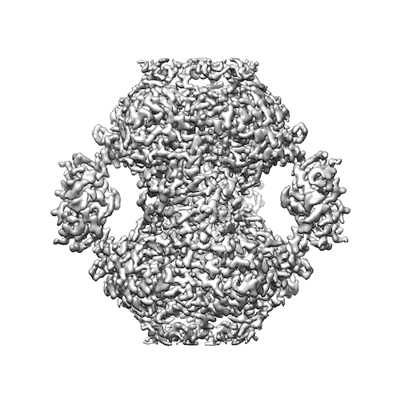
HUMAN IMPDH1 TREATED WITH ATP, IMP, AND NAD+
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 7rer
Q-score: 0.675
Burrell AL, Nie C, Said M, Simonet JC, Fernandez-Justel D, Johnson MC, Quispe J, Buey RM, Peterson JR, Kollman JM
Nat Struct Mol Biol (2022) 29 pp. 47-58 [ DOI: doi:10.1038/s41594-021-00706-2 Pubmed: 35013599 ]
- Assembly interface of impdh1 filament bound to atp, imp, nad+ (55 GDa, Complex from Homo sapiens)
- Inosinic acid (348 Da, Ligand)
- Isoform 5 of inosine-5'-monophosphate dehydrogenase 1 (55 kDa, Protein from Homo sapiens)
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
HUMAN IMPDH1 TREATED WITH ATP, IMP, AND NAD+; OCTAMER-CENTERED
Resolution: 3.05 Å
EM Method: Single-particle
Fitted PDBs: 7res
Q-score: 0.575
Burrell AL, Nie C, Said M, Simonet JC, Fernandez-Justel D, Johnson MC, Quispe J, Buey RM, Peterson JR, Kollman JM
Nat Struct Mol Biol (2022) 29 pp. 47-58 [ DOI: doi:10.1038/s41594-021-00706-2 Pubmed: 35013599 ]
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Assembly interface of impdh1 filament bound to atp, imp, nad+ (55 GDa, Complex from Homo sapiens)
- Inosinic acid (348 Da, Ligand)
- Isoform 5 of inosine-5'-monophosphate dehydrogenase 1 (55 kDa, Protein from Homo sapiens)
HUMAN IMPDH1 TREATED WITH GTP, IMP, AND NAD+; INTERFACE-CENTERED
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 7rfe
Q-score: 0.64
Burrell AL, Nie C, Said M, Simonet JC, Fernandez-Justel D, Johnson MC, Quispe J, Buey RM, Peterson JR, Kollman JM
Nat Struct Mol Biol (2022) 29 pp. 47-58 [ DOI: doi:10.1038/s41594-021-00706-2 Pubmed: 35013599 ]
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Assembly interface of impdh1 filament bound to atp, imp, nad+ (55 GDa, Complex from Homo sapiens)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Inosine-5'-monophosphate dehydrogenase 1 (54 kDa, Protein from Homo sapiens)
- Inosinic acid (348 Da, Ligand)
HUMAN IMPDH1 TREATED WITH GTP, IMP, AND NAD+ OCTAMER-CENTERED
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 7rfg
Q-score: 0.649
Burrell AL, Nie C, Said M, Simonet JC, Fernandez-Justel D, Johnson MC, Quispe J, Buey RM, Peterson JR, Kollman JM
Nat Struct Mol Biol (2022) 29 pp. 47-58 [ DOI: doi:10.1038/s41594-021-00706-2 Pubmed: 35013599 ]
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Assembly interface of impdh1 filament bound to atp, imp, nad+ (55 GDa, Complex from Homo sapiens)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Inosinic acid (348 Da, Ligand)
- Isoform 5 of inosine-5'-monophosphate dehydrogenase 1 (55 kDa, Protein from Homo sapiens)
HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH GTP, ATP, IMP, NAD+; INTERFACE-CENTERED
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7rgi
Q-score: 0.491
Burrell AL, Nie C, Said M, Simonet JC, Fernandez-Justel D, Johnson MC, Quispe J, Buey RM, Peterson JR, Kollman JM
Nat Struct Mol Biol (2022) 29 pp. 47-58 [ DOI: doi:10.1038/s41594-021-00706-2 Pubmed: 35013599 ]
- Assembly interface of impdh1 filament bound to atp, imp, nad+ (55 GDa, Complex from Homo sapiens)
- Inosinic acid (348 Da, Ligand)
- Inosine-5'-monophosphate dehydrogenase 1 (58 kDa, Protein from Homo sapiens)
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH ATP, IMP, NAD+, INTERFACE-CENTERED
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 7rgl
Q-score: 0.673
Burrell AL, Nie C, Said M, Simonet JC, Fernandez-Justel D, Johnson MC, Quispe J, Buey RM, Peterson JR, Kollman JM
Nat Struct Mol Biol (2022) 29 pp. 47-58 [ DOI: doi:10.1038/s41594-021-00706-2 Pubmed: 35013599 ]
- Assembly interface of impdh1 filament bound to atp, imp, nad+ (55 GDa, Complex from Homo sapiens)
- Inosine-5'-monophosphate dehydrogenase 1 (58 kDa, Protein from Homo sapiens)
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Inosinic acid (348 Da, Ligand)
HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH ATP, IMP, NAD+, OCTAMER-CENTERED
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 7rgm
Q-score: 0.587
Burrell AL, Nie C, Said M, Simonet JC, Fernandez-Justel D, Johnson MC, Quispe J, Buey RM, Peterson JR, Kollman JM
Nat Struct Mol Biol (2022) 29 pp. 47-58 [ DOI: doi:10.1038/s41594-021-00706-2 Pubmed: 35013599 ]
- Inosine-5'-monophosphate dehydrogenase 1 (58 kDa, Protein from Homo sapiens)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Assembly interface of impdh1 filament bound to atp, imp, nad+ (55 GDa, Complex from Homo sapiens)
- Inosinic acid (348 Da, Ligand)
HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH GTP, ATP, IMP, NAD+; INTERFACE-CENTERED
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 7rgq
Q-score: 0.502
Burrell AL, Nie C, Said M, Simonet JC, Fernandez-Justel D, Johnson MC, Quispe J, Buey RM, Peterson JR, Kollman JM
Nat Struct Mol Biol (2022) 29 pp. 47-58 [ DOI: doi:10.1038/s41594-021-00706-2 Pubmed: 35013599 ]
- Inosine-5'-monophosphate dehydrogenase 1 (58 kDa, Protein from Homo sapiens)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Assembly interface of impdh1 filament bound to atp, imp, nad+ (55 GDa, Complex from Homo sapiens)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Inosinic acid (348 Da, Ligand)
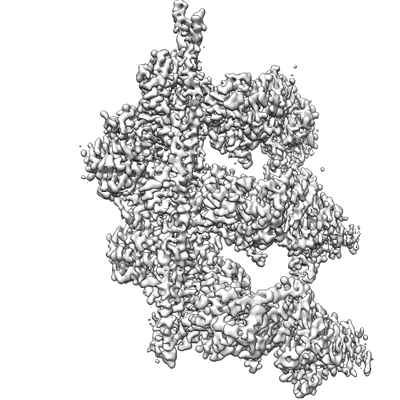
Cryo-EM structure of BCL6 bound to BI-3802
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 6xmx
Q-score: 0.285
Slabicki M, Yoon H, Koeppel J, Nitsch L, Roy Burman SS, Di Genua C, Donovan KA, Sperling AS, Hunkeler M, Tsai JM, Sharma R, Guirguis A, Zou C, Chudasama P, Gasser JA, Miller PG, Scholl C, Frohling S, Nowak RP, Fischer ES, Ebert BL
Nature (2020) 588 pp. 164-168 [ Pubmed: 33208943 DOI: doi:10.1038/s41586-020-2925-1 ]
- B-cell lymphoma 6 protein (bcl6) filament (352 kDa, Cellular component from Homo sapiens)
- B-cell lymphoma 6 protein (44 kDa, Protein from Homo sapiens)
- 2-[6-[[5-chloranyl-2-[(3~{s},5~{r})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{n}-methyl-ethanamide (484 Da, Ligand)
Resolution: 3.8 Å
EM Method: Helical reconstruction
Fitted PDBs: 7jh7
Q-score: 0.308
Risi C, Schafer LU, Belknap B, Pepper I, White HD, Schroder GF, Galkin VE
Structure (2021) 29 pp. 50- [ Pubmed: 33065066 DOI: doi:10.1016/j.str.2020.09.013 ]
- Porcine native cardiac thin filament decorated with porcine cardiac myosin-s1 (492 MDa, Complex from Sus scrofa)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Actin, alpha cardiac muscle 1 (42 kDa, Protein from Sus scrofa)
- Tropomyosin (11 kDa, Protein from Sus scrofa)
- Myosin-7 (223 kDa, Protein from Sus scrofa)