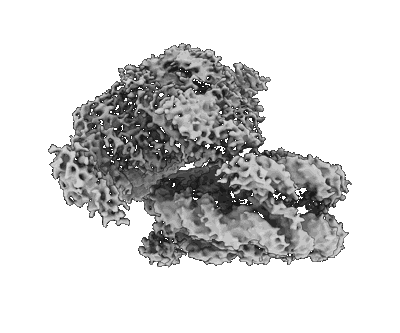
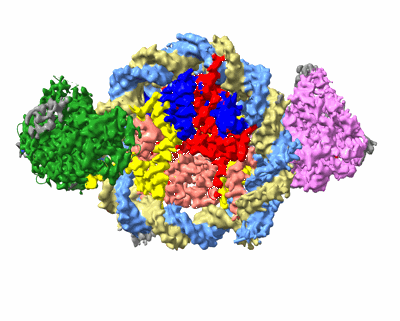
Cryo-EM structure of native SWR1 bound to nucleosome (composite structure)
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 9b1e
Q-score: 0.191
Louder RK, Park G, Ye Z, Cha JS, Gardner AM, Lei Q, Ranjan A, Hollmuller E, Stengel F, Pugh BF, Wu C
Cell (2024) 187 pp. 6849-6864.e18 [ Pubmed: 39357520 DOI: doi:10.1016/j.cell.2024.09.007 ]
- Dna (214-mer) (66 kDa, DNA from synthetic construct)
- Vacuolar protein sorting-associated protein 72 (90 kDa, Protein from Saccharomyces cerevisiae W303)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Ruvb-like protein 1 (91 kDa, Protein from Saccharomyces cerevisiae W303)
- Histone h2a (14 kDa, Protein from Saccharomyces cerevisiae)
- Dna (214-mer) (65 kDa, DNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Native swr1 bound to nucleosome. (1 MDa, Complex from Saccharomyces cerevisiae)
- Helicase swr1 (178 kDa, Protein from Saccharomyces cerevisiae W303)
- Histone h4 (11 kDa, Protein from Drosophila melanogaster)
- Zinc ion (65 Da, Ligand)
- Actin-like protein arp6 (50 kDa, Protein from Saccharomyces cerevisiae W303)
- Beryllium trifluoride ion (66 Da, Ligand)
- Swr1-complex protein 3 (72 kDa, Protein from Saccharomyces cerevisiae W303)
- Vacuolar protein sorting-associated protein 71 (32 kDa, Protein from Saccharomyces cerevisiae W303)
- Ruvb-like protein 2 (51 kDa, Protein from Saccharomyces cerevisiae W303)
- Histone h3 (15 kDa, Protein from Drosophila melanogaster)
- Native swr1 complex (Complex from Saccharomyces cerevisiae)
- Histone h2b (14 kDa, Protein from Saccharomyces cerevisiae)
SWR1-nucleosome complex in configuration 2
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 8qkv
Q-score: 0.185
Girvan P, Jalal ASB, McCormack EA, Skehan MT, Knight CL, Wigley DB, Rueda DS
Nature (2024) 636 pp. 251-257 [ Pubmed: 39506114 DOI: doi:10.1038/s41586-024-08152-y ]
- Magnesium ion (24 Da, Ligand)
- Histone h2a.2 (17 kDa, Protein from Saccharomyces cerevisiae S288C)
- Vacuolar protein sorting-associated protein 71 (32 kDa, Protein from Saccharomyces cerevisiae S288C)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Helicase swr1 (174 kDa, Protein from Saccharomyces cerevisiae S288C)
- Histone h3 (15 kDa, Protein from Saccharomyces cerevisiae S288C)
- Beryllium trifluoride ion (66 Da, Ligand)
- Actin-like protein arp6 (50 kDa, Protein from Saccharomyces cerevisiae S288C)
- Zinc ion (65 Da, Ligand)
- Swr1-nucleosome complex (Complex from Saccharomyces cerevisiae)
- Dna (194-mer) (60 kDa, DNA from synthetic construct)
- Vacuolar protein sorting-associated protein 72 (21 kDa, Protein from Saccharomyces cerevisiae S288C)
- Histone h2b.1 (14 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ruvb-like protein 1 (50 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ruvb-like protein 2 (51 kDa, Protein from Saccharomyces cerevisiae S288C)
- Histone h4 (11 kDa, Protein from Saccharomyces cerevisiae S288C)
- Dna (194-mer) (59 kDa, DNA from synthetic construct)
SWR1 lacking Swc5 subunit in complex with hexasome
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 9fbw
Q-score: 0.193
Jalal ASB, Girvan P, Chua EYD, Liu L, Wang S, McCormack EA, Skehan MT, Knight CL, Rueda DS, Wigley DB
Mol Cell (2024) 84 pp. 3871- [ Pubmed: 39226902 DOI: doi:10.1016/j.molcel.2024.08.015 ]
- Magnesium ion (24 Da, Ligand)
- Vacuolar protein sorting-associated protein 71 (32 kDa, Protein from Saccharomyces cerevisiae S288C)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Helicase swr1 (174 kDa, Protein from Saccharomyces cerevisiae S288C)
- Histone h3 (15 kDa, Protein from Saccharomyces cerevisiae S288C)
- Dna (112-mer) (34 kDa, DNA from synthetic construct)
- Dna (112-mer) (34 kDa, DNA from synthetic construct)
- Actin-like protein arp6 (50 kDa, Protein from Saccharomyces cerevisiae S288C)
- Beryllium trifluoride ion (66 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Histone h2b.1 (14 kDa, Protein from Saccharomyces cerevisiae S288C)
- Vacuolar protein sorting-associated protein 72 (90 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ruvb-like protein 1 (50 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ruvb-like protein 2 (51 kDa, Protein from Saccharomyces cerevisiae S288C)
- Histone h4 (11 kDa, Protein from Saccharomyces cerevisiae S288C)
- Swr1 lacking swc5 subunit in complex with hexasome (Complex from Saccharomyces cerevisiae S288C)
- Histone h2a.1 (14 kDa, Protein from Saccharomyces cerevisiae S288C)
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 9ipu
Q-score: 0.237
Shi Q, Deng ZH, Zhang LY, Tong ZB, Chu G-C, Ai HS, Liu L
To Be Published
- Ubiquitin (8 kDa, Protein from Homo sapiens)
- Histone h3.2 (15 kDa, Protein from Homo sapiens)
- Dna (171-mer) (52 kDa, DNA from Homo sapiens)
- Ubiquitin-conjugating enzyme e2 d3 (16 kDa, Protein from Homo sapiens)
- E3 ubiquitin-protein ligase rnf168 (22 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Histone h2a type 1-b/e (14 kDa, Protein from Homo sapiens)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Histone h2b type 1-k (13 kDa, Protein from Homo sapiens)
- Dna (171-mer) (52 kDa, DNA from Homo sapiens)
- Histone h1.0 (20 kDa, Protein from Homo sapiens)
- Cryo-em structure of the rnf168(1-193)/ubch5c-ub ubiquitylation module bound to h1.0-k63-ub3 modified chromatosome (Complex from Homo sapiens)
PRC2 monomer bound to nucleosome
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 8tas
Q-score: 0.259
Sauer PV, Pavlenko E, Cookis T, Zirden LC, Renn J, Singhal A, Hunold P, Hoehne-Wiechmann MN, van Ray O, Kaschani F, Kaiser M, Hansel-Hertsch R, Sanbonmatsu KY, Nogales E, Poepsel S
Mol Cell (2024) 84 pp. 3885-3898.e8 [ DOI: doi:10.1016/j.molcel.2024.08.025 Pubmed: 39303719 ]
- Polycomb protein suz12 (72 kDa, Protein from Homo sapiens)
- Histone h2a (14 kDa, Protein from Xenopus laevis)
- Zinc finger protein aebp2 (34 kDa, Protein from Homo sapiens)
- Polycomb protein eed (50 kDa, Protein from Homo sapiens)
- Prc2 monomer bound to nucleosome (290 kDa, Complex from Homo sapiens)
- Histone-lysine n-methyltransferase ezh2 (86 kDa, Protein from Homo sapiens)
- Dna (226-mer) (66 kDa, DNA from synthetic construct)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Dna (226-mer) (65 kDa, DNA from synthetic construct)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Prc2 (Complex from Homo sapiens)
- Histone-binding protein rbbp4 (47 kDa, Protein from Homo sapiens)
- Nucleosome (Complex from Xenopus laevis)
PRC2-J119-450 monomer bound to H1-nucleosome
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 8tb9
Q-score: 0.261
Sauer PV, Pavlenko E, Cookis T, Zirden LC, Renn J, Singhal A, Hunold P, Hoehne-Wiechmann MN, van Ray O, Kaschani F, Kaiser M, Hansel-Hertsch R, Sanbonmatsu KY, Nogales E, Poepsel S
Mol Cell (2024) 84 pp. 3885-3898.e8 [ DOI: doi:10.1016/j.molcel.2024.08.025 Pubmed: 39303719 ]
- Histone h2a type 1 (14 kDa, Protein from Xenopus laevis)
- Histone h1.0 (21 kDa, Protein from Homo sapiens)
- Polycomb protein suz12 (72 kDa, Protein from Homo sapiens)
- Zinc finger protein aebp2 (34 kDa, Protein from Homo sapiens)
- Polycomb protein eed (50 kDa, Protein from Homo sapiens)
- Prc2-j119-450 monomer bound to h1-nucleosome (290 kDa, Complex from Homo sapiens)
- Histone-lysine n-methyltransferase ezh2 (86 kDa, Protein from Homo sapiens)
- Dna (226-mer) (66 kDa, DNA from synthetic construct)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Protein jumonji (36 kDa, Protein from Homo sapiens)
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Dna (226-mer) (65 kDa, DNA from synthetic construct)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Prc2 (Complex from Homo sapiens)
- Histone-binding protein rbbp4 (47 kDa, Protein from Homo sapiens)
- Nucleosome (Complex from Xenopus laevis)
Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp.
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 9gd1
Q-score: 0.332
Engeholm M, Roske JJ, Oberbeckmann E, Dienemann C, Lidschreiber M, Cramer P, Farnung L
Mol Cell (2024) 84 pp. 3423- [ DOI: doi:10.1016/j.molcel.2024.08.022 Pubmed: 39270644 ]
- Dna (261-mer) (81 kDa, DNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromo domain-containing protein 1 (168 kDa, Protein from Saccharomyces cerevisiae S288C)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Beryllium trifluoride ion (66 Da, Ligand)
- Histone h2a type 1 (14 kDa, Protein from Xenopus laevis)
- Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp (Complex from Saccharomyces cerevisiae S288C)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Dna (261-mer) (80 kDa, DNA from synthetic construct)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp.
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 9gd2
Q-score: 0.295
Engeholm M, Roske JJ, Oberbeckmann E, Dienemann C, Lidschreiber M, Cramer P, Farnung L
Mol Cell (2024) 84 pp. 3423- [ DOI: doi:10.1016/j.molcel.2024.08.022 Pubmed: 39270644 ]
- Magnesium ion (24 Da, Ligand)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromo domain-containing protein 1 (168 kDa, Protein from Saccharomyces cerevisiae S288C)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Beryllium trifluoride ion (66 Da, Ligand)
- Dna (248-mer) (77 kDa, DNA from synthetic construct)
- Histone h2a type 1 (14 kDa, Protein from Xenopus laevis)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. (Complex from Xenopus laevis)
- Dna (248-mer) (76 kDa, DNA from synthetic construct)
Cryo-EM structure of the Rpd3S-nucleosome complex from budding yeast in State 3
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 8w9f
Q-score: 0.156
Sci Adv (2024) 10 pp. eadk7678-eadk7678 [ DOI: doi:10.1126/sciadv.adk7678 Pubmed: 38598631 ]
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Histone h2a type 1-b/e (14 kDa, Protein from Homo sapiens)
- Transcriptional regulatory protein rco1 (78 kDa, Protein from Saccharomyces cerevisiae)
- Histone h3.1 (15 kDa, Protein from Homo sapiens)
- Transcriptional regulatory protein sin3 (175 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- The rpd3s complex (Complex from Saccharomyces cerevisiae)
- Histone h2b type 1-k (13 kDa, Protein from Homo sapiens)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- 3-dna (45 kDa, DNA from Homo sapiens)
- 5-dna (45 kDa, DNA from Homo sapiens)
Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2
Resolution: 4.05 Å
EM Method: Single-particle
Fitted PDBs: 8wha
Q-score: 0.306
Liu Y, Zhang Z, Hu H, Chen W, Zhang F, Wang Q, Wang C, Yan K, Du J
Nat Plants (2024) 10 pp. 374-380 [ Pubmed: 38413824 DOI: doi:10.1038/s41477-024-01640-z ]
- Dna (sense strand) (45 kDa, DNA from synthetic construct)
- Dna (antisense strand) (45 kDa, DNA from synthetic construct)
- Beryllium trifluoride ion (66 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Histone h4 (11 kDa, Protein from Arabidopsis thaliana)
- Histone h3.1 (15 kDa, Protein from Arabidopsis thaliana)
- Histone h2a.6 (13 kDa, Protein from Arabidopsis thaliana)
- Ddm1-nucleosome complex in the adp-befx state with ddm1 bound to shl2 and shl-2 (376 kDa, Complex from Arabidopsis thaliana)
- Atp-dependent dna helicase ddm1 (86 kDa, Protein from Arabidopsis thaliana)
- Histone h2b.6 (16 kDa, Protein from Arabidopsis thaliana)