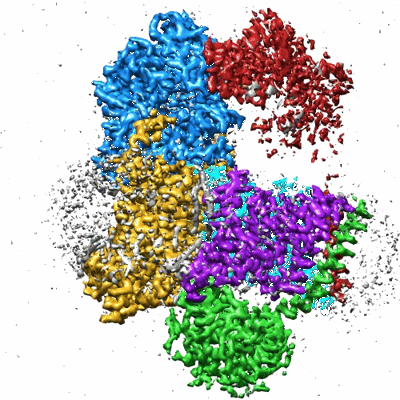
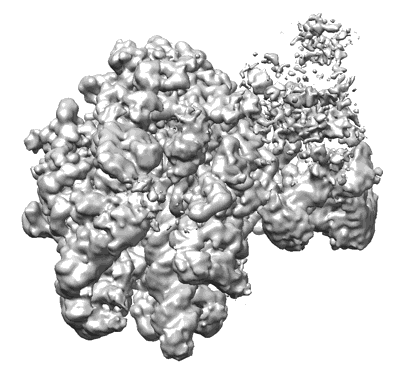
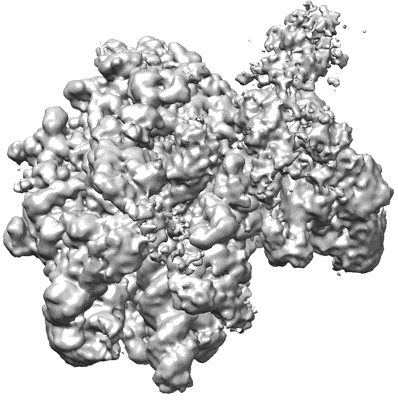
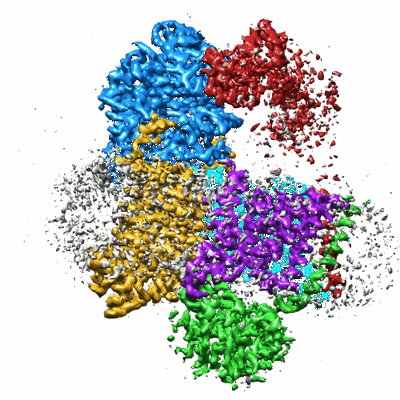
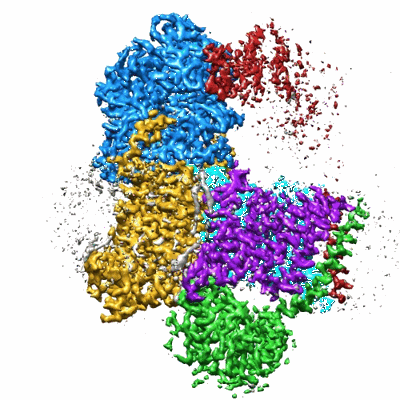
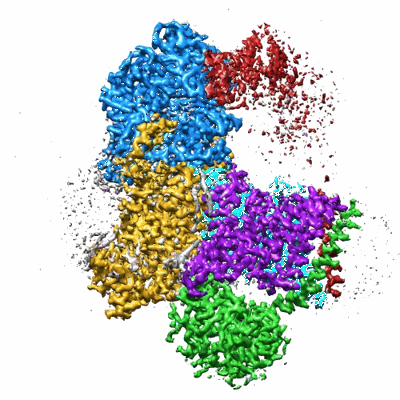
Sodium pumping NADH-quinone oxidoreductase with substrates NADH and Q2
Resolution: 2.86 Å
EM Method: Single-particle
Fitted PDBs: 8a1u
Q-score: 0.558
Hau JL, Kaltwasser S, Muras V, Casutt MS, Vohl G, Claussen B, Steffen W, Leitner A, Bill E, Cutsail 3rd GE, DeBeer S, Vonck J, Steuber J, Fritz G
Nat Struct Mol Biol (2023) 30 pp. 1686-1694 [ DOI: doi:10.1038/s41594-023-01099-0 Pubmed: 37710014 ]
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae)
- Ubiquinone-2 (318 Da, Ligand)
- 1,4-dihydronicotinamide adenine dinucleotide (665 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae)
- Water (18 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit a (51 kDa, Protein from Vibrio cholerae)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae)
- Sodium ion (22 Da, Ligand)
- Nqr complex with substrates nadh and q2 (220 kDa, Complex from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)
Structural insights into human co-transcriptional capping - structure 1
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8p4a
Q-score: 0.344
Garg G, Dienemann C, Farnung L, Schwarz J, Linden A, Urlaub H, Cramer P
Mol Cell (2023) 83 pp. 2464-2477.e5 [ Pubmed: 37369200 DOI: doi:10.1016/j.molcel.2023.06.002 ]
- Dna-directed rna polymerase ii subunit rpb3 (31 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb11-a (13 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase subunit beta (134 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit e (24 kDa, Protein from Sus scrofa)
- Magnesium ion (24 Da, Ligand)
- Dna-directed rna polymerase ii subunit f (14 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (17 kDa, Protein from Sus scrofa)
- Dna (29-mer) (8 kDa, DNA from unidentified)
- Dna-directed rna polymerase ii subunit rpb7 (19 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb9 (14 kDa, Protein from Sus scrofa)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerase subunit (218 kDa, Protein from Sus scrofa)
- Rna polymerase ii subunit k (7 kDa, Protein from Sus scrofa)
- Pol ii - tpase complex (Complex from Homo sapiens)
- Mrna-capping enzyme (68 kDa, Protein from Homo sapiens)
- Rna polymerase ii subunit d (16 kDa, Protein from Sus scrofa)
- Rna (5'-r(p*ap*ap*ap*ap*cp*cp*cp*ap*gp*gp*gp*ap*ap*cp*cp*cp*ap*cp*u)-3') (6 kDa, RNA from unidentified)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (7 kDa, Protein from Sus scrofa)
- Dna (38-mer) (11 kDa, DNA from unidentified)
Structural insights into human co-transcriptional capping - structure 2
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8p4b
Q-score: 0.434
Garg G, Dienemann C, Farnung L, Schwarz J, Linden A, Urlaub H, Cramer P
Mol Cell (2023) 83 pp. 2464-2477.e5 [ Pubmed: 37369200 DOI: doi:10.1016/j.molcel.2023.06.002 ]
- Dna-directed rna polymerase ii subunit rpb3 (31 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb11-a (13 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase subunit beta (134 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit e (24 kDa, Protein from Sus scrofa)
- Magnesium ion (24 Da, Ligand)
- Dna-directed rna polymerase ii subunit f (14 kDa, Protein from Sus scrofa)
- Mrna-capping enzyme (68 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (17 kDa, Protein from Sus scrofa)
- Dna (35-mer) (10 kDa, DNA from unidentified)
- Dna-directed rna polymerase ii subunit rpb7 (19 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb1 (217 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb9 (14 kDa, Protein from Sus scrofa)
- Rna (5'-r(p*gp*up*ap*ap*cp*cp*gp*gp*ap*gp*ap*gp*gp*gp*ap*ap*cp*cp*cp*ap*cp*u)-3') (7 kDa, RNA from unidentified)
- Zinc ion (65 Da, Ligand)
- Rna polymerase ii subunit k (7 kDa, Protein from Sus scrofa)
- Pol ii - tpase complex (Complex from Homo sapiens)
- Dna (5'-d(p*ap*gp*cp*gp*gp*cp*gp*gp*ap*gp*cp*cp*ap*gp*cp*ap*gp*gp*gp*ap*gp*cp*tp*g)-3') (7 kDa, DNA from unidentified)
- Rna polymerase ii subunit d (16 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (7 kDa, Protein from Sus scrofa)
Structural insights into human co-transcriptional capping - structure 3
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 8p4c
Q-score: 0.297
Garg G, Dienemann C, Farnung L, Schwarz J, Linden A, Urlaub H, Cramer P
Mol Cell (2023) 83 pp. 2464-2477.e5 [ Pubmed: 37369200 DOI: doi:10.1016/j.molcel.2023.06.002 ]
- Dna-directed rna polymerase ii subunit rpb3 (31 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb11-a (13 kDa, Protein from Sus scrofa)
- Transcription elongation factor spt4 (13 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase subunit beta (134 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit e (24 kDa, Protein from Sus scrofa)
- Magnesium ion (24 Da, Ligand)
- Dna (41-mer) (12 kDa, DNA from unidentified)
- Mrna-capping enzyme (68 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase ii subunit f (14 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (17 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb7 (19 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb1 (217 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb9 (14 kDa, Protein from Sus scrofa)
- Zinc ion (65 Da, Ligand)
- Rna polymerase ii subunit k (7 kDa, Protein from Sus scrofa)
- Pol ii - tpase complex (Complex from Homo sapiens)
- Transcription elongation factor spt5 (121 kDa, Protein from Homo sapiens)
- Rna polymerase ii subunit d (16 kDa, Protein from Sus scrofa)
- Rna (5'-r(p*cp*gp*gp*ap*gp*ap*gp*gp*gp*ap*ap*cp*cp*cp*ap*cp*u)-3') (5 kDa, RNA from unidentified)
- Dna (32-mer) (9 kDa, DNA from unidentified)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (7 kDa, Protein from Sus scrofa)
Structural insights into human co-transcriptional capping - structure 4
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8p4d
Q-score: 0.348
Garg G, Dienemann C, Farnung L, Schwarz J, Linden A, Urlaub H, Cramer P
Mol Cell (2023) 83 pp. 2464-2477.e5 [ Pubmed: 37369200 DOI: doi:10.1016/j.molcel.2023.06.002 ]
- Dna-directed rna polymerase ii subunit rpb3 (31 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb11-a (13 kDa, Protein from Sus scrofa)
- Transcription elongation factor spt4 (13 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase subunit beta (134 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit e (24 kDa, Protein from Sus scrofa)
- Rna (5'-r(p*ap*cp*cp*gp*gp*ap*gp*ap*gp*gp*gp*ap*ap*cp*cp*cp*ap*cp*u)-3') (6 kDa, RNA from unidentified)
- Dna (41-mer) (12 kDa, DNA from unidentified)
- Dna-directed rna polymerase ii subunit f (14 kDa, Protein from Sus scrofa)
- Mrna-capping enzyme (68 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (17 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb7 (19 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb1 (217 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb9 (14 kDa, Protein from Sus scrofa)
- Zinc ion (65 Da, Ligand)
- Rna polymerase ii subunit k (7 kDa, Protein from Sus scrofa)
- Pol ii - tpase complex (Complex from Homo sapiens)
- Transcription elongation factor spt5 (121 kDa, Protein from Homo sapiens)
- Rna polymerase ii subunit d (16 kDa, Protein from Sus scrofa)
- Dna (32-mer) (9 kDa, DNA from unidentified)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (7 kDa, Protein from Sus scrofa)
Structural insights into human co-transcriptional capping - structure 5
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 8p4e
Q-score: 0.297
Garg G, Dienemann C, Farnung L, Schwarz J, Linden A, Urlaub H, Cramer P
Mol Cell (2023) 83 pp. 2464- [ Pubmed: 37369200 DOI: doi:10.1016/j.molcel.2023.06.002 ]
- Dna-directed rna polymerase ii subunit rpb3 (31 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb11-a (13 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase subunit beta (134 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit e (24 kDa, Protein from Sus scrofa)
- Cap-specific mrna (nucleoside-2'-o-)-methyltransferase 1 (95 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Dna-directed rna polymerase ii subunit f (14 kDa, Protein from Sus scrofa)
- Mrna-capping enzyme (68 kDa, Protein from Homo sapiens)
- Rna (5'-d(*(mgt))-r(p*gp*ap*cp*ap*up*ap*cp*ap*up*ap*ap*ap*gp*ap*cp*cp*ap*gp*gp*c)-3') (6 kDa, RNA from synthetic construct)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (17 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb7 (19 kDa, Protein from Sus scrofa)
- Dna-directed rna polymerase ii subunit rpb1 (217 kDa, Protein from Sus scrofa)
- Dna (26-mer) (8 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase ii subunit rpb9 (14 kDa, Protein from Sus scrofa)
- Zinc ion (65 Da, Ligand)
- Rna polymerase ii subunit k (7 kDa, Protein from Sus scrofa)
- Pol ii - tpase complex (Complex from Homo sapiens)
- Transcription elongation factor spt5 (121 kDa, Protein from Homo sapiens)
- Rna polymerase ii subunit d (16 kDa, Protein from Sus scrofa)
- Dna (35-mer) (10 kDa, DNA from synthetic construct)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (7 kDa, Protein from Sus scrofa)
Sodium pumping NADH-quinone oxidoreductase
Resolution: 3.37 Å
EM Method: Single-particle
Fitted PDBs: 8a1t
Q-score: 0.452
Hau JL, Kaltwasser S, Muras V, Casutt MS, Vohl G, Claussen B, Steffen W, Leitner A, Bill E, Cutsail 3rd GE, DeBeer S, Vonck J, Steuber J, Fritz G
Nat Struct Mol Biol (2023) 30 pp. 1686-1694 [ DOI: doi:10.1038/s41594-023-01099-0 Pubmed: 37710014 ]
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae)
- Nqr complex (220 kDa, Complex from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Potassium ion (39 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae)
- Sodium ion (22 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit a (48 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)
Sodium pumping NADH-quinone oxidoreductase with substrate Q2
Resolution: 2.73 Å
EM Method: Single-particle
Fitted PDBs: 8a1v
Q-score: 0.54
Hau JL, Kaltwasser S, Muras V, Casutt MS, Vohl G, Claussen B, Steffen W, Leitner A, Bill E, Cutsail 3rd GE, DeBeer S, Vonck J, Steuber J, Fritz G
Nat Struct Mol Biol (2023) 30 pp. 1686-1694 [ DOI: doi:10.1038/s41594-023-01099-0 Pubmed: 37710014 ]
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae)
- Ubiquinone-2 (318 Da, Ligand)
- Water (18 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit a (51 kDa, Protein from Vibrio cholerae)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae)
- Sodium ion (22 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Nqr complex with substrate q2 (220 kDa, Complex from Vibrio cholerae)
- Flavin mononucleotide (456 Da, Ligand)
Sodium pumping NADH-quinone oxidoreductase with substrate Q1
Resolution: 2.56 Å
EM Method: Single-particle
Fitted PDBs: 8a1w
Q-score: 0.555
Hau JL, Kaltwasser S, Muras V, Casutt MS, Vohl G, Claussen B, Steffen W, Leitner A, Bill E, Cutsail 3rd GE, DeBeer S, Vonck J, Steuber J, Fritz G
Nat Struct Mol Biol (2023) 30 pp. 1686-1694 [ DOI: doi:10.1038/s41594-023-01099-0 Pubmed: 37710014 ]
- Nqr complex with substrate q1 (220 kDa, Complex from Vibrio cholerae)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae)
- Ubiquinone-1 (250 Da, Ligand)
- Water (18 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit a (51 kDa, Protein from Vibrio cholerae)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae)
- Sodium ion (22 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)
Sodium pumping NADH-quinone oxidoreductase with inhibitor DQA
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8a1x
Q-score: 0.467
Hau JL, Kaltwasser S, Muras V, Casutt MS, Vohl G, Claussen B, Steffen W, Leitner A, Bill E, Cutsail 3rd GE, DeBeer S, Vonck J, Steuber J, Fritz G
Nat Struct Mol Biol (2023) 30 pp. 1686-1694 [ DOI: doi:10.1038/s41594-023-01099-0 Pubmed: 37710014 ]
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- Nqr complex with inhibitor dqa (220 kDa, Complex from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit a (48 kDa, Protein from Vibrio cholerae)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)