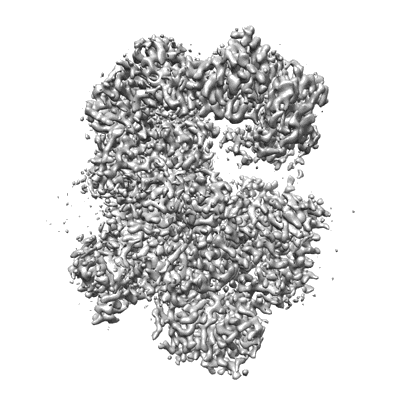
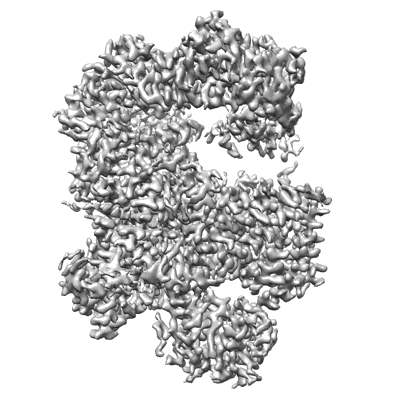
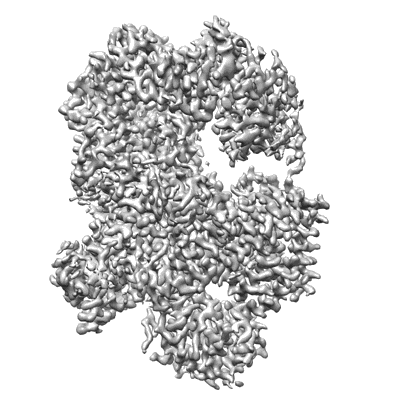
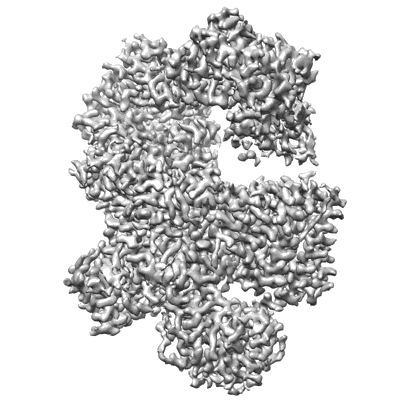
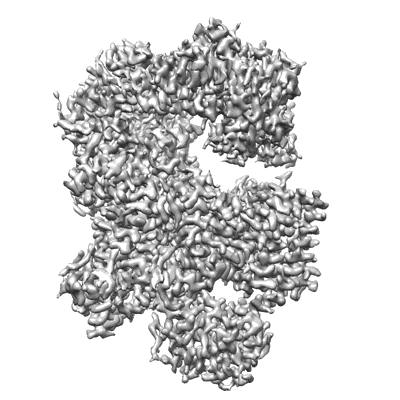Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 1
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7xk3
Q-score: 0.567
Kishikawa JI, Ishikawa M, Masuya T, Murai M, Kitazumi Y, Butler NL, Kato T, Barquera B, Miyoshi H
Nat Commun (2022) 13 pp. 4082-4082 [ Pubmed: 35882843 DOI: doi:10.1038/s41467-022-31718-1 ]
- Na(+)-translocating nadh-quinone reductase subunit a (48 kDa, Protein from Vibrio cholerae O395)
- Calcium ion (40 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (744 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae O395)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae O395)
- Na+-pumping nadh-ubiquinone oxidoreductase from vibrio cholerae, state 1 (220 kDa, Complex from Vibrio cholerae O395)
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae O395)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae O395)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae O395)
Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 2
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7xk4
Q-score: 0.562
Kishikawa JI, Ishikawa M, Masuya T, Murai M, Kitazumi Y, Butler NL, Kato T, Barquera B, Miyoshi H
Nat Commun (2022) 13 pp. 4082-4082 [ Pubmed: 35882843 DOI: doi:10.1038/s41467-022-31718-1 ]
- Na(+)-translocating nadh-quinone reductase subunit a (48 kDa, Protein from Vibrio cholerae O395)
- Calcium ion (40 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (744 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae O395)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae O395)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae O395)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae O395)
- Na+-pumping nadh-ubiquinone oxidoreductase from vibrio cholerae, state 2 (220 kDa, Complex from Vibrio cholerae O395)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae O395)
Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 3
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7xk5
Q-score: 0.546
Kishikawa JI, Ishikawa M, Masuya T, Murai M, Kitazumi Y, Butler NL, Kato T, Barquera B, Miyoshi H
Nat Commun (2022) 13 pp. 4082-4082 [ Pubmed: 35882843 DOI: doi:10.1038/s41467-022-31718-1 ]
- Na(+)-translocating nadh-quinone reductase subunit a (48 kDa, Protein from Vibrio cholerae O395)
- Na+-pumping nadh-ubiquinone oxidoreductase from vibrio cholerae, state 3 (220 kDa, Complex from Vibrio cholerae O395)
- Calcium ion (40 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (744 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae O395)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae O395)
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae O395)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae O395)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae O395)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7xk6
Q-score: 0.575
Kishikawa JI, Ishikawa M, Masuya T, Murai M, Kitazumi Y, Butler NL, Kato T, Barquera B, Miyoshi H
Nat Commun (2022) 13 pp. 4082-4082 [ Pubmed: 35882843 DOI: doi:10.1038/s41467-022-31718-1 ]
- Na(+)-translocating nadh-quinone reductase subunit a (48 kDa, Protein from Vibrio cholerae O395)
- Water (18 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (744 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae O395)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae O395)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae O395)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Aurachin d (363 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae O395)
- Na+-pumping nadh-ubiquinone oxidoreductase from vibrio cholerae, with aurachin d-42 (220 kDa, Complex from Vibrio cholerae O395)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae O395)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 7xk7
Q-score: 0.584
Kishikawa JI, Ishikawa M, Masuya T, Murai M, Kitazumi Y, Butler NL, Kato T, Barquera B, Miyoshi H
Nat Commun (2022) 13 pp. 4082-4082 [ Pubmed: 35882843 DOI: doi:10.1038/s41467-022-31718-1 ]
- Na(+)-translocating nadh-quinone reductase subunit a (48 kDa, Protein from Vibrio cholerae O395)
- Water (18 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Flavin mononucleotide (456 Da, Ligand)
- Riboflavin (376 Da, Ligand)
- Korormicin (433 Da, Ligand)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (744 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit c (27 kDa, Protein from Vibrio cholerae O395)
- Na(+)-translocating nadh-quinone reductase subunit e (21 kDa, Protein from Vibrio cholerae O395)
- Na(+)-translocating nadh-quinone reductase subunit b (45 kDa, Protein from Vibrio cholerae O395)
- Na+-pumping nadh-ubiquinone oxidoreductase from vibrio cholerae, with korormicin (220 kDa, Complex from Vibrio cholerae O395)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit f (45 kDa, Protein from Vibrio cholerae O395)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Na(+)-translocating nadh-quinone reductase subunit d (22 kDa, Protein from Vibrio cholerae O395)
V/A-type ATPase/synthase from Thermus thermophilus, peripheral domain, rotational state 1
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 5y5y
Q-score: 0.362
Nakanishi A, Kishikawa JI, Tamakoshi M, Mitsuoka K, Yokoyama K
Nat Commun (2018) 9 pp. 89-89 [ DOI: doi:10.1038/s41467-017-02553-6 Pubmed: 29311594 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- V-type atp synthase subunit e (20 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase beta chain (53 kDa, Protein from Thermus thermophilus HB8)
- V/a-type atpase/synthase from thermus thermophilus, peripheral domai, rotational state 1. (650 kDa, Complex from Thermus thermophilus HB8)
- V-type atp synthase subunit c (35 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit f (11 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit d (24 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase, subunit (vapc-therm) (13 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase alpha chain (63 kDa, Protein from Thermus thermophilus HB8)
V/A-type ATPase/synthase from Thermus thermophilus, rotational state 1
Resolution: 5.0 Å
EM Method: Single-particle
Fitted PDBs: 5y5x
Q-score: 0.303
Nakanishi A, Kishikawa JI, Tamakoshi M, Mitsuoka K, Yokoyama K
Nat Commun (2018) 9 pp. 89-89 [ DOI: doi:10.1038/s41467-017-02553-6 Pubmed: 29311594 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- V-type atp synthase subunit e (20 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase beta chain (53 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit c (35 kDa, Protein from Thermus thermophilus HB8)
- V/a-type atpase/synthase from thermus thermophilus, rotational state 1. (650 kDa, Complex from Thermus thermophilus HB8)
- V-type atp synthase subunit f (11 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit d (24 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase, subunit (vapc-therm) (13 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit i (72 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase alpha chain (63 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase, subunit k (9 kDa, Protein from Thermus thermophilus HB8)
V/A-type ATPase/synthase from Thermus thermophilus, rotational state 2
Resolution: 6.7 Å
EM Method: Single-particle
Fitted PDBs: 5y5z
Q-score: 0.237
Nakanishi A, Kishikawa JI, Tamakoshi M, Mitsuoka K, Yokoyama K
Nat Commun (2018) 9 pp. 89-89 [ DOI: doi:10.1038/s41467-017-02553-6 Pubmed: 29311594 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- V-type atp synthase subunit e (20 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase beta chain (53 kDa, Protein from Thermus thermophilus HB8)
- V/a-type atpase/synthase from thermus thermophilus, rotational state 2. (650 kDa, Complex from Thermus thermophilus HB8)
- V-type atp synthase subunit c (35 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit f (11 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit d (24 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase, subunit (vapc-therm) (13 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit i (72 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase alpha chain (63 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase, subunit k (9 kDa, Protein from Thermus thermophilus HB8)
V/A-type ATPase/synthase from Thermus thermophilus, rotational state 3.
Resolution: 7.5 Å
EM Method: Single-particle
Fitted PDBs: 5y60
Q-score: 0.232
Nakanishi A, Kishikawa JI, Tamakoshi M, Mitsuoka K, Yokoyama K
Nat Commun (2018) 9 pp. 89-89 [ DOI: doi:10.1038/s41467-017-02553-6 Pubmed: 29311594 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- V-type atp synthase subunit e (20 kDa, Protein from Thermus thermophilus HB8)
- V/a-type atpase/synthase from thermus thermophilus, rotational state 3. (650 kDa, Complex from Thermus thermophilus HB8)
- V-type atp synthase beta chain (53 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit c (35 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit f (11 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit d (24 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase, subunit (vapc-therm) (13 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase subunit i (72 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase alpha chain (63 kDa, Protein from Thermus thermophilus HB8)
- V-type atp synthase, subunit k (9 kDa, Protein from Thermus thermophilus HB8)