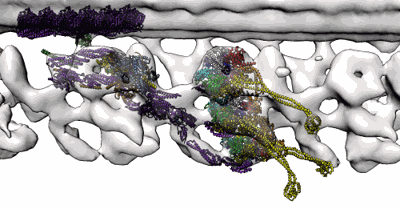
In situ outer dynein arm from Chlamydomonas reinhardtii in a pre-power stroke state
Resolution: 38.0 Å
EM Method: Subtomogram averaging
Fitted PDBs: 8bwy
Q-score: 0.036
Zimmermann N, Noga A, Obbineni JM, Ishikawa T
EMBO J (2023) 42 pp. e112466-e112466 [ Pubmed: 37051721 DOI: doi:10.15252/embj.2022112466 ]
- Docking complex 1/2 protein (11 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein-1-alpha heavy chain, flagellar inner arm i1 complex protein, putative (475 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein light chain tctex-type 1 protein (13 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein light chain (13 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein light chain roadblock-type 2 protein (18 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein light chain roadblock-type 2 protein (14 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein light chain (10 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein light chain (10 kDa, Protein from Chlamydomonas reinhardtii)
- In situ outer dynein arm (Cellular component from Chlamydomonas reinhardtii)
- Dynein light chain (10 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein light chain 2a (15 kDa, Protein from Chlamydomonas reinhardtii)
- Flagellar outer dynein arm intermediate protein, putative (77 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein light chain (12 kDa, Protein from Chlamydomonas reinhardtii)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Dynein heavy chain, outer arm protein (534 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein intermediate chain 2 (77 kDa, Protein from Chlamydomonas reinhardtii)
- Thioredoxin (12 kDa, Protein from Chlamydomonas reinhardtii)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Calmodulin (35 kDa, Protein from Chlamydomonas reinhardtii)
- Dynein light chain (12 kDa, Protein from Chlamydomonas reinhardtii)
- Outer arm dynein beta heavy chain (530 kDa, Protein from Chlamydomonas reinhardtii)
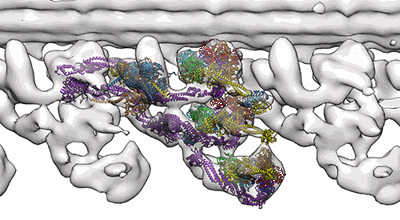
In situ outer dynein arm from Chlamydomonas reinhardtii in the post-power stroke state
Resolution: 30.3 Å
EM Method: Subtomogram averaging
Fitted PDBs: 8bx8
Q-score: 0.043
Zimmermann N, Noga A, Obbineni JM, Ishikawa T
EMBO J (2023) 42 pp. e112466-e112466 [ DOI: doi:10.15252/embj.2022112466 Pubmed: 37051721 ]
- Thioredoxin (14 kDa, Protein from Tetrahymena thermophila)
- Dynein intermediate chain 2 (76 kDa, Protein from Tetrahymena thermophila)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Dynein light chain 1 (22 kDa, Protein from Tetrahymena thermophila)
- Dynein light chain 4a (17 kDa, Protein from Tetrahymena thermophila)
- Dynein light chain roadblock-type 2 protein (18 kDa, Protein from Tetrahymena thermophila)
- Dynein light chain tctex-type 1 protein (13 kDa, Protein from Tetrahymena thermophila)
- Flagellar outer dynein arm intermediate protein, putative (77 kDa, Protein from Tetrahymena thermophila)
- Dynein light chain (11 kDa, Protein from Tetrahymena thermophila)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Dynein light chain 2a (15 kDa, Protein from Tetrahymena thermophila)
- Dynein light chain roadblock-type 2 protein (14 kDa, Protein from Tetrahymena thermophila)
- Dynein light chain (12 kDa, Protein from Tetrahymena thermophila)
- Magnesium ion (24 Da, Ligand)
- Dynein heavy chain, outer arm protein (534 kDa, Protein from Tetrahymena thermophila)
- Outer arm dynein beta heavy chain (530 kDa, Protein from Tetrahymena thermophila)
- In situ outer dynein arm (Cellular component from Chlamydomonas reinhardtii)
- Dynein light chain (10 kDa, Protein from Tetrahymena thermophila)
- Dynein light chain (12 kDa, Protein from Tetrahymena thermophila)
- Dynein-1-alpha heavy chain, flagellar inner arm i1 complex protein, putative (475 kDa, Protein from Tetrahymena thermophila)
- Dynein light chain (10 kDa, Protein from Tetrahymena thermophila)
- Dynein light chain (10 kDa, Protein from Tetrahymena thermophila)
COPII on membranes, outer coat right-handed rod, class1
Resolution: 40.0 Å
EM Method: Subtomogram averaging
Fitted PDBs: 6zg5
Q-score: 0.067
Hutchings J, Stancheva VG, Brown NR, Cheung ACM, Miller EA, Zanetti G
Nat Commun (2021) 12 pp. 2034-2034 [ Pubmed: 33795673 DOI: doi:10.1038/s41467-021-22110-6 ]
- Protein transport protein sec31 (138 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Protein transport protein sec13 (33 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Protein transport protein sec13 (33 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Copii coat assembled on lipid bilayer (Complex from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Water (18 Da, Ligand)
Volume 1 for truncated dimeric Cytohesin-3 (Grp1; amino acids 14-399)
Resolution: 53.0 Å
EM Method: Single-particle
Fitted PDBs: 6u3e
Q-score: 0.043
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
Structure (2019) 27 pp. 1782- [ Pubmed: 31601460 DOI: doi:10.1016/j.str.2019.09.007 ]
Resolution: 53.0 Å
EM Method: Single-particle
Fitted PDBs: 6u3g
Q-score: 0.048
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
Structure (2019) 27 pp. 1782- [ Pubmed: 31601460 DOI: doi:10.1016/j.str.2019.09.007 ]
Resolution: 35.0 Å
EM Method: Single-particle
Fitted PDBs: 6bbp
Q-score: 0.056
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
Structure (2018) 26 pp. 106-117.e6 [ DOI: doi:10.1016/j.str.2017.11.019 Pubmed: 29276036 ]
- Magnesium ion (24 Da, Ligand)
- Cytohesin-3,adp-ribosylation factor 6 (60 kDa, Protein from Homo sapiens)
- Truncated monomeric cytohesin-3 (grp1; amino acids 63-399) e161a 6gs arf6 q67l fusion protein complex with gtp, mg and inositol 1,3,4,5 tetrakisphosphate (Complex from Mus musculus)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Inositol-(1,3,4,5)-tetrakisphosphate (500 Da, Ligand)
Resolution: 35.0 Å
EM Method: Single-particle
Fitted PDBs: 6bbq
Q-score: 0.057
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
Structure (2018) 26 pp. 106-117.e6 [ Pubmed: 29276036 DOI: doi:10.1016/j.str.2017.11.019 ]
- Magnesium ion (24 Da, Ligand)
- Inositol-(1,3,4,5)-tetrakisphosphate (500 Da, Ligand)
- Cytohesin-3,adp-ribosylation factor 6 (60 kDa, Protein from Homo sapiens)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Truncated monomeric cytohesin-3 (grp1; amino acids 63-399) e161a 6gs arf6 q67l fusion protein (Complex from Mus musculus)
Subtomogram-averaged map of a single rubella virus capsid unit
Resolution: 35.0 Å
EM Method: Subtomogram averaging
Fitted PDBs: 5khe
Q-score: 0.07
Mangala Prasad V, Klose T, Rossmann MG
PLoS Pathog (2017) 13 pp. e1006377-e1006377 [ Pubmed: 28575072 DOI: doi:10.1371/journal.ppat.1006377 ]
- Water (18 Da, Ligand)
- Rubella virus strain m33 (64 kDa, Complex from Rubella virus strain M33)
- Capsid protein (30 kDa, Protein from Rubella virus)
Subtomogram-averaged map of a single rubella virus capsid unit
Resolution: 35.0 Å
EM Method: Subtomogram averaging
Fitted PDBs: 5khf
Q-score: 0.062
Mangala Prasad V, Klose T, Rossmann MG
PLoS Pathog (2017) 13 pp. e1006377-e1006377 [ Pubmed: 28575072 DOI: doi:10.1371/journal.ppat.1006377 ]
- Water (18 Da, Ligand)
- Rubella virus strain m33 (64 kDa, Complex from Rubella virus strain M33)
- Capsid protein (30 kDa, Protein from Rubella virus)